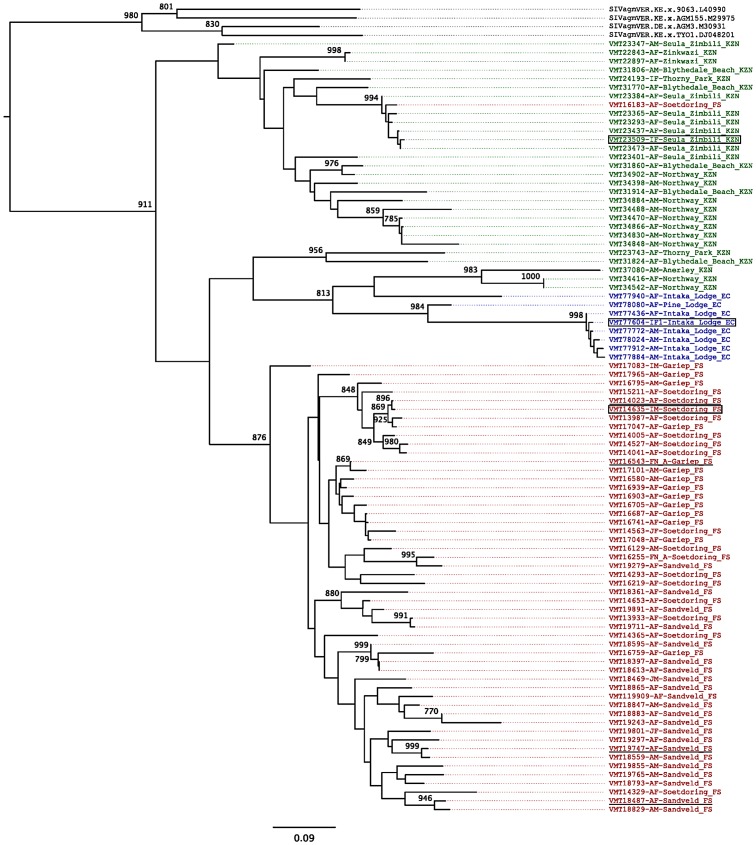
Figure 2. SIVagmVer diversity in wild AGMs from different parks and game reserves in the Republic of South Africa.
Maximum likelihood tree for pol gene. Sequences are colored depending on the region in which the sequences were sampled, with red indicating sequences from Free State, green indicating sequences from KwaZulu-Natal, and blue indicating sequences from Eastern Cape territories. Reference SIVagm strains from vervet monkeys in Kenya are shown in black. The tree is based on a 592 bp pol sequence alignment (after gap-containing sites were removed). Maximum likelihood estimates were performed using 1000 replicate bootstrap analysis. Internal nodes indicate level of support values for internal branching. Bar represents number of amino acid replacements (0.09) per site. The strain nomenclature includes the identification number, monkey status (M-male, F-female; I-infant; J-juvenile and A-adult),and the site and state of origin (FS-Free State; KZN-KwaZulu Natal; EC-Eastern Cape). Putative heterosexual transmission cases documented to be of recent occurrence based on VL and serology testing are underlined. Putative maternal-to-infant transmission cases are boxed.