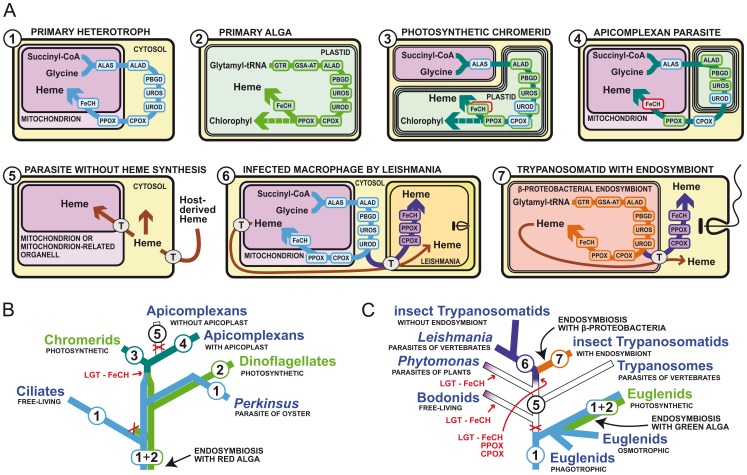
Figure 1. Different ways of heme acquisition and their distributions within alveolates and kinetoplastids.
(A) Intracellular localizations of heme synthesis enzymes and heme trafficking in various parasites. The colors and numbers of the phylogenetic lineages in (B) and (C) correspond to the colors of the pathways and numbers in the schematics in (A). The blue line (no. 1) represents the original pathway of the primary heterotroph and is similar to the well characterized C4 pathways of animals and fungi. The green line (no. 2) represents the plastidial C5 pathway of the endosymbiotic alga and is present in most phototrophs. In the ancestral lineages of alveolates, both of these pathways co-existed as in extant photosynthetic euglenids. The lines no. 3 and 4 represent the hybrid pathways of photosynthetic chromerids and apicomplexan parasites, which resulted from the combination of the original pathway of the host (no. 1) and the plastidial pathway from the algal endosymbiont (no. 2). FeCH in a red box was acquired via lateral gene transfer (LGT) from a γ-proteobacterium. The purple line (no. 6) represents the partial pathway of the trypanosomatid flagellates, which consists of the last three steps encoded by enzymes of γ-proteobacterial origin. The orange line (no. 7) is the heme synthesis pathway of the β-proteobacterial endosymbionts of one group of trypanosomatids. T stands for porphyrin transporters. (B) Evolution of the heme biosynthesis in Alveolata leading to the unique pathway of apicomplexan parasites. (C) Evolution of the heme synthesis in Euglenozoa leading to the loss of the pathway in ancestral kinetoplastids and its partial rescue by bacterial genes for the last three steps and full rescue by bacterial endosymbionts in trypanosomatids. The abbreviated enzyme names are: ALAS, ALA-synthase; GTR, glutamyl-tRNA reductase; GSA-AT, glutamate 1-semialdehyde aminotransferase; ALAD, ALA-dehydratase; PBGD, porphobilinogen deaminase; UROS, uroporphyrinogen synthase; UROD, uroporphyrinogen decarboxylase; CPOX, coproporphyrinogen oxidase; PPOX, protoporphyrinogen oxidase; FeCH, ferrochelatase.