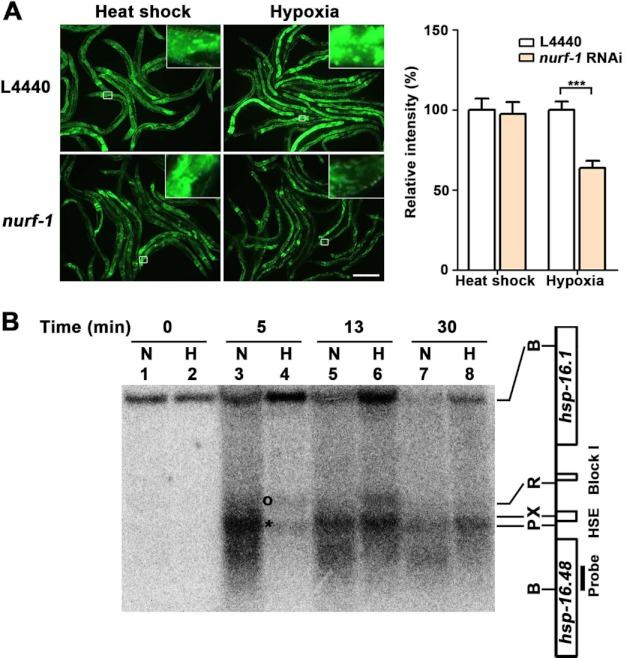
FIGURE 3.
NURF-1 chromatin-remodeling factor mediates the hypoxia response of the hsp-16.1 gene. A, decreased nurf-1 function (Drosophila NURF301) reduced the activation of hsp-16.1 by hypoxia. The results for ACF and CHRAC RNAi are presented in supplemental Fig. S7. Quantification of the relative intensity of HSP-16.1::GFP fluorescence was carried out using ImageJ software. Statistical significance was determined using an unpaired t test (***, p < 0.001). Error bars indicate S.E. Scale bar = 200 μm. B, MNase digestion patterns of the normoxic (N) and hypoxic (H) chromatins. The DNA of MNase-digested nuclei was digested by BglII and analyzed by Southern blot hybridization using a 200-nucleotide probe made from the region designated in the figure. Chromatin was incubated in the presence of MNase for 0 min (lanes 1 and 2), 5 min (lanes 3 and 4), 13 min (lanes 5 and 6), and 30 min (lanes 7 and 8). The asterisk indicates the genomic position that is more sensitive to MNase cleavage in normoxia. The circle indicates the band with increased sensitivity to MNase cleavage in hypoxia. B, BglII; P, PstI; X, XbaI; R, RsaI; HSE, heat shock response element.