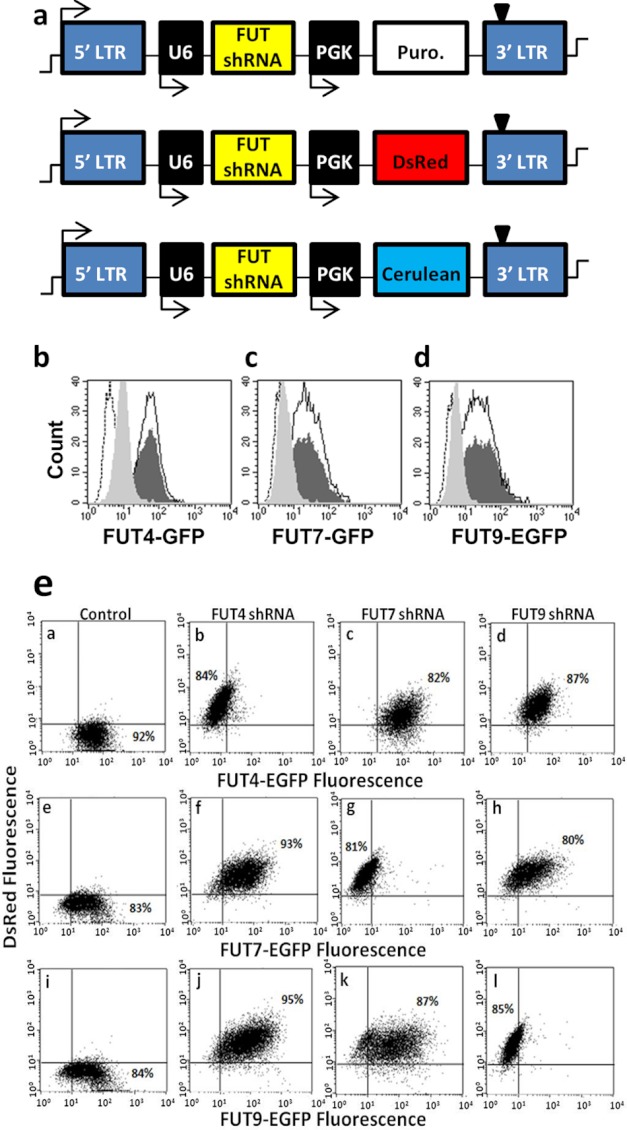
FIGURE 3.
Determination of specific and efficient shRNA that knocks down FUTs. a, pLKO.1 plasmids contain the polymerase-III U6 promoter that drives shRNA expression. The PGK promoter drives either the puromycin (Puro.) resistance gene or reporter genes (DsRed or cerulean). b–d, flow cytometry histogram showing fluorescence of CHO-S cells with either FUT4-EGFP (b), FUT7-EGFP (c), or FUT9-EGFP (d). Fluorescence decreases in each panel from dark-shaded histogram for cells without shRNA to light-shaded histogram upon gene silencing. Empty histogram with black border corresponds to TRC control virus, whereas dashed-empty histogram corresponds to WT CHO-S cells lacking FUT-EGFP. Silencing efficiency was quantified based on % reduction in EGFP fluorescence intensity upon shRNA addition in comparison with control FUT-EGFP cell fluorescence. e, two-color cross-silencing cytometry experiments were performed to confirm that each shRNA only disrupts a unique fucosyltransferase. To this end, CHO-S cells that stably express FUT4-EGFP (top row), FUT7-EGFP (middle row), and FUT9-EGFP (bottom row) were either left untransduced (1st column) or these were transduced with virus encoding for shRNA against FUT4 (2nd column), FUT7 (3rd column), or FUT9 (last column). All shRNA viruses carried the DsRed gene in addition to shRNA, and thus transduction results in an increase in DsRed signal in all cases. Silencing specificity is evident because each shRNA only reduced the EGFP fluorescence of corresponding FUT-EGFP cell lines.