FIGURE 5.
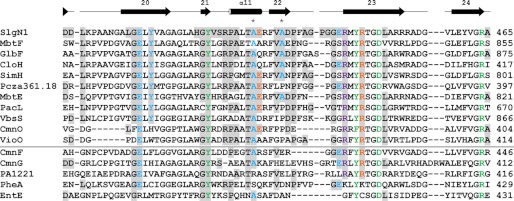
Sequence alignment of the Acore interface region of SlgN1. The alignment is divided by a horizontal line separating MbtH-dependent (top) and MbtH-independent (bottom) A-domains. The last residue of the sequences is numbered on the right. Additionally, Ala-428 and Ala-433 of SlgN1 are marked with an asterisk. Residues that are involved in interface formation in SlgN1 are highlighted (gray) and colored by their type of interaction as follows: blue, hydrogen bonding; orange, salt bridges; violet, π-stacking interaction; green, active site residues. The secondary structure assigned at the top corresponds to SlgN1. The alignment for MbtH-dependent and -independent A-domains was generated separately using the program ClustalW2 (51) and a Gonnet matrix. MbtH-dependent proteins include MbtF and MbtE (9), GlbF (6), CloH, SimH, and Pcza361.18 (11), PacL and VbsS (8), and CmnO and VioO (7). MbtH-independent proteins include CmnF and CmnG (7), PA1221 (52), PheA (11, 28), and EntE (7, 53).
