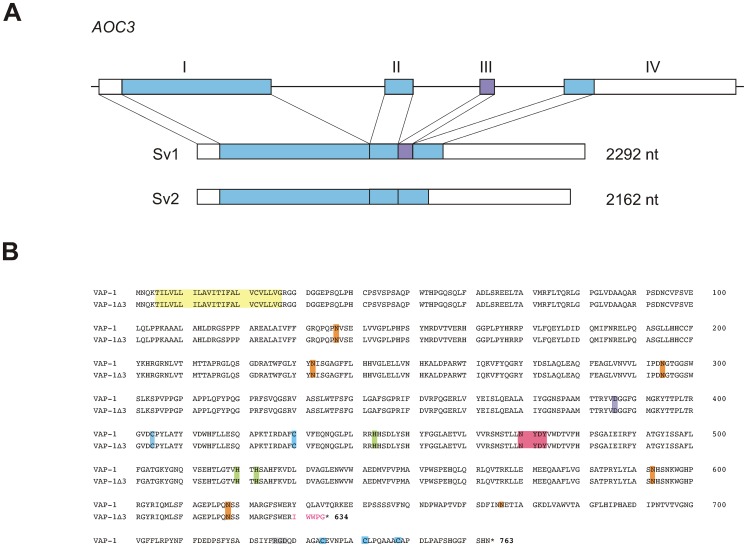
Figure 1. Characteristics of the alternatively spliced transcript of VAP-1.
(A) Schematic presentation of the exon–intron organization of the human VAP-1 gene, AOC3. The boxes with roman numerals (I-IV) represent the exons. The translated regions are shown in color and the 5′- and 3′-untranslated regions in white. Exon III (violet) is spliced out in the shorter splice variant. Sv1: the full-length splice variant; Sv2: the alternatively spliced shorter splice variant. (B) Sequence alignment of the two VAP-1 isoforms. The deduced amino acid sequences of VAP-1 and the shorter isoform VAP-1Δ3. Highlighted are: light yellow, the hydrophobic N-terminal sequence; pink, the conserved signature motif of the active site, in which the first tyrosine is post-translationally modified to topaquinone; lilac, the (putative) catalytic site base; light green, the conserved Cu(II) binding histidine residues; light blue, the conserved cysteine residues involved in dimerization; orange the putative N-linked glycosylation sites, grey, RGD sequence. The amino acids unique to VAP-1Δ3 are in red.