Figure 2.
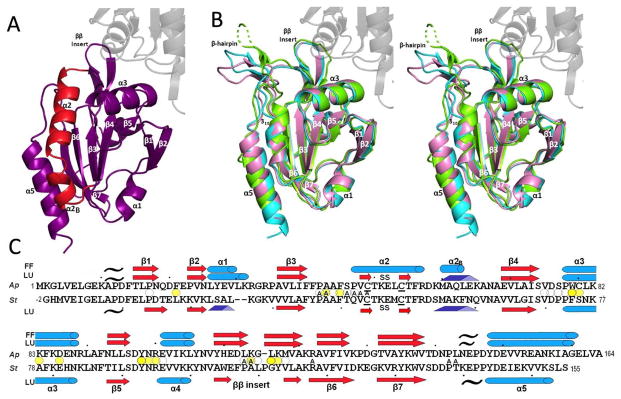
Overall structures of ApPrxQ and StPrxQ. (A) The FF subunit of the ApPrxQ-FF/LU structure is shown with the main segment involved in the conformational transition (residues 46–67) highlighted in red. The universally conserved Prx secondary structures are labeled (except α4 which is behind β5). (B) Representative LU chains are superimposed for ApPrxQ-FF/LU (violet), ApPrxQ-LU (cyan), and StPrxQ (green). The un-modeled part of the StPrxQ β-hairpin is shown as a dashed line. The area of transition highlighted in panel A becomes a β-hairpin and 310 helix (both labeled). For perspective, here and in panel A the second subunit of the dimer is shown for ApPrxQ-LU (ghost gray). (C) Structure-based sequence alignment of ApPrxQ and StPrxQ indicating placements of α-helices (cyan cylinders), β-strands (red arrows and pale red arrows for the inferred StPrxQ β-hairpin), 310 helices (blue triangular rods), and PII spirals (black tildes). Ellipsoids mark residues with surface area buried at the dimer interface of >100 Å2 (dark yellow), 25 to 100 Å2 (medium yellow) and <25 Å2 (white). ApPrxQ residues having surface area buried by the modeled DTT inhibitor are noted with A for Active-Site.
