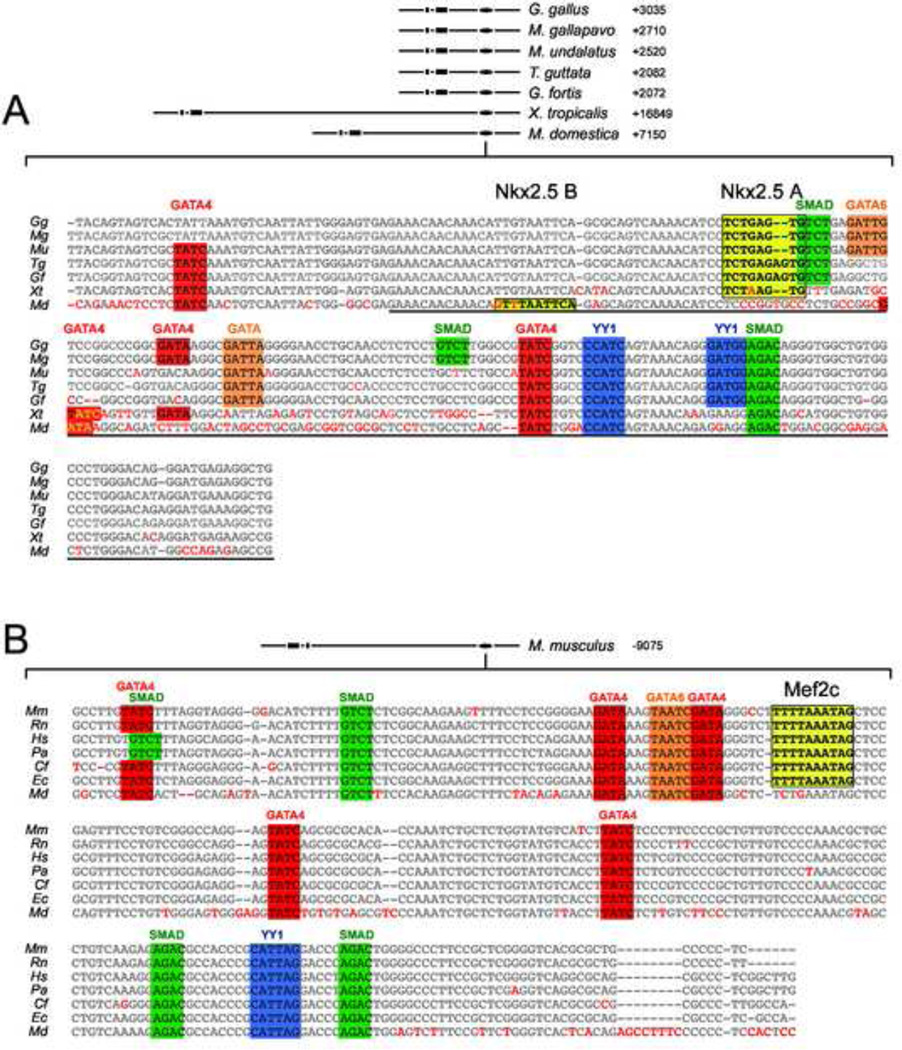
Figure 3. Divergent conserved enhancers mediate SHF expression of Nkx2.5 in mammalian and non-mammalian vertebrates.
Shown in A is the central BMP response element contained with the chicken Gallus gallus (Gg) Nkx2.5 SHF enhancer (CAR3) aligned with homologous 3’ flanking regions in the Nkx2.5 genes from the turkey Meleagris gallopavo (Mg), Budgarigar or parakeet Melopsittacus undalatus (Mu), medium ground finch Geospiza fortis (Gf), zebrafinch Taenopygia guttata (Tg), frog Xenopustropicalis (Xt) and opossum Monodelphis domestica (Md). Homologous flanking regions are at varying distances 3’ from the transcriptional start site in these species, as shown in the schematic at top. Conserved residues are shown in gray and divergent residues are highlighted in red. Consensus binding motifs are shown as colored boxes for cardiac GATA (red), Smad4 (green), YY1 (blue) and Nkx2.5 (yellow). Underlined region highlights the previously characterized chicken Nkx2.5 BMPRE. B. SHF enhancer conserved in mammalian species. Shown are aligned Nkx2.5-5’ enhancer sequences from mouse Mus musculus (Mm), rat Rattus norvegicus (Rn), human Homo sapiens (Hs), chimp Pan troglodytes (Pt), orangutan Pongo pygmaetus abelii (Pa), dog Canis familiaris (Cf), horse Equus caballus (Ec), and opossum Monodelphis domestica (Md). TF site highlighting is as in A, except that a conserved Mef2 CArG binding consensus rather than an NKE is in yellow. Note that both enhancer motifs are conserved in opossum, but lack either a strong Nkx2.5 binding motif (3’ enhancer) or Mef2 binding CArG consensus (5’ enhancer).