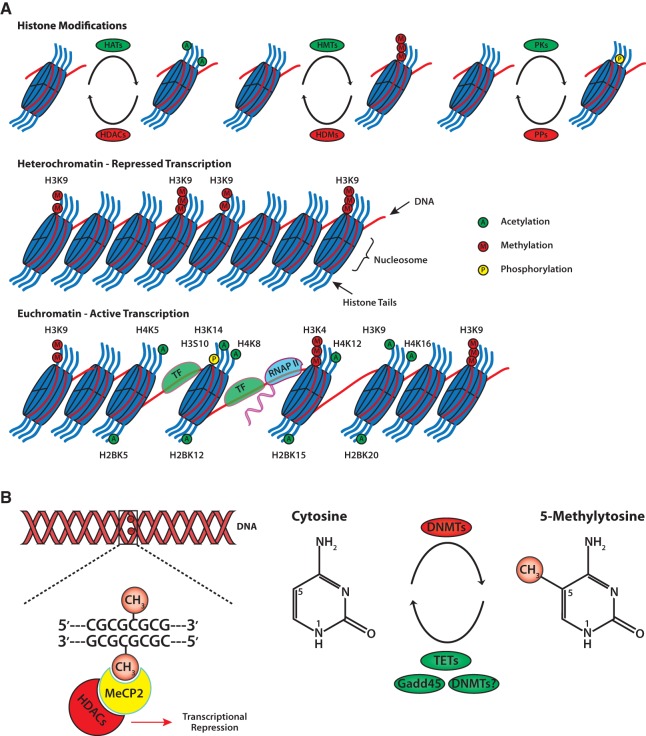
Figure 1.
General schematic of epigenetic modifications. (A) Packaging of DNA into chromatin is achieved through the wrapping of 146 bp of DNA around octamers of histone proteins. Chromatin-modifying enzymes dynamically regulate the addition and removal of post-translational modifications on histone N-terminal tails. Modifications associated with learning and memory include histone acetylation, phosphorylation, and methylation. The specific combination of histone tail modifications dictate whether or not the chromatin exists as heterochromatin or euchromatin. Heterochromatin is characterized by condensed chromatin and subsequent transcriptional repression. Euchromatin is characterized by a relaxed chromatin state that allows transcriptional machinery access to DNA for gene expression. (B) Methylation of DNA involves covalent addition of a methyl group to the 5′ position of the cytosine pyrimidine ring by DNMTs. DNA methylation commonly occurs at genes enriched with cytosine-guanine nucleotides (CpG islands). Proteins with methyl-binding domains, like MeCP2, bind to methylated DNA and recruit repressor complexes containing HDACs. Recent evidence suggests that active DNA demethylation can occur via several mechanisms involving members of the Gadd45 family, TET family, and DNMTs themselves. (DNMTs) DNA methyltransferases, (Gadd45) growth arrest and DNA damage 45, (HATs) histone acetyltransferases, (HDACs) histone deacetylases, (HDMs) histone demethylases, (HMTs) histone methyltransferases, (MeCP2) methyl CpG binding protein 2, (PKs) protein kinases, (PPs) protein phosphatases, (TETs) ten eleven translocation, (TF) transcription factor, (RNAP II) RNA polymerase II.