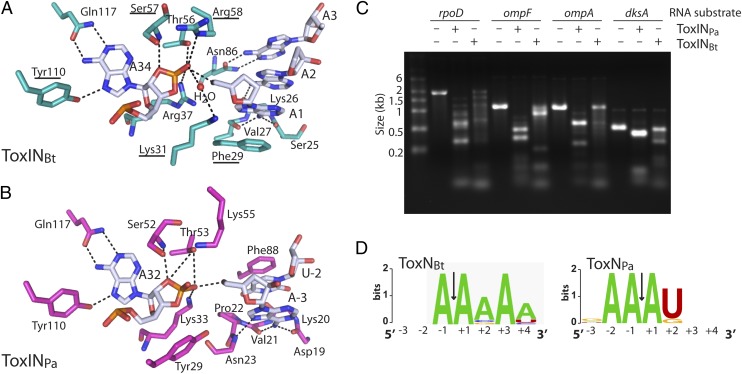
Fig. 8.
ToxNBt and ToxNPa are endoribonucleases with selective substrate preferences. (A) View of ToxINBt active site showing key interactions for product recognition and substrate cleavage. ToxNBt is shown in teal and ToxIBt RNA in white. Hydrogen bonds are indicated by black dashed lines. Residues that were mutated resulting in a loss of toxicity are underlined (see also Fig. 7C). Phosphate groups of the ToxIBt backbone are omitted for clarity. (B) Active site of ToxINPa (PDB ID 2XDB). ToxNPa is shown in light magenta and ToxIPa in white. (C) In vitro degradation products of ToxINPa and ToxINBt. Substrate RNAs were incubated with purified ToxIN complex at a 1:4 molar ratio, and the products were examined by agarose gel electrophoresis. (D) Sequence-preference profiles of ToxNBt and ToxNPa, generated by performing 5′ RACE on the ToxINBt- and ToxINPa-cleavage products of E. coli rpoD, ompF, ompA, and dksA RNAs. The sequence-preference profiles shown are from a total of 14 (ToxINPa) and 12 (ToxINBt) unique 5′ ends.