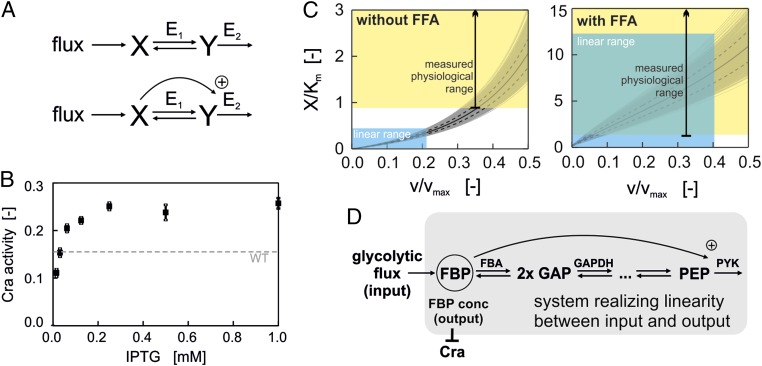
Fig. 2.
(A) (Upper) Structure of mathematical model without feedforward activation (FFA). E1 is assumed to follow reversible Michaelis–Menten kinetics, and E2 is assumed to follow irreversible Michaelis–Menten kinetics. (Lower) Structure of mathematical model with FFA of E2 by X. E1 is assumed to follow reversible Michaelis–Menten kinetics, and E2 is assumed to follow MWC kinetics. (B) Cra activity as a function of IPTG concentration (used as a proxy for pyruvate kinase abundance) determined in glucose batch cultures of a pykF mutant strain bearing an IPTG-inducible PYK I expression plasmid (black squares). Dashed line, Cra activity in wild-type strain in glucose batch culture. (C) Simulation of model without (Left) or with (Right) FFA. Kinetic parameters: Km = Km.X.E1 = Km.Y.E1 = Km.Y.E2 = 0.2 mM; vmax = vmax.E1 = vmax.E2 = 1 mM/s; Keq = 50; KmA.X.E2 = 0.6 mM; L = 4·106; n = 4. KmA.X.E2, L, and n were used only for the model including FFA. The gray lines show the range of X when repeating the simulation 1,000 times while sampling the parameter values from a uniform distribution within 10% deviation of the original parameter values. The continuous black lines show the mean value of X across all simulations, and the dashed black lines show the corresponding SD. The blue areas are visual aids to highlight the approximate linear range of X. The yellow areas denote the range of physiological X/Km ratios. The green shading indicates the area, where a linear relationship between X and v is possible at physiological X/Km ranges. This is only the case for the system with the feedforward activation. (D) Structure of the flux sensing mechanism: Reversible reactions between FBP and PEP couple FBP to lower glycolysis, and the FFA of PYK by FBP is essential for establishing the linear correlation of FBP and glycolytic flux beyond the Km value.