Abstract
In recent years, studies of zebrafish rod and cone photoreceptors have yielded novel insights into the differentiation of distinct photoreceptor cell types and the mechanisms guiding photoreceptor regeneration following cell death, and they have provided models of human retinal degeneration. These studies were facilitated by the use of transgenic zebrafish expressing fluorescent reporter genes under the control of various cell-specific promoters. Improvements in transgenesis techniques (e.g., Tol2 transposition), the availability of numerous fluorescent reporter genes with different localization properties, and the ability to generate transgenes via recombineering (e.g., Gateway technology) have enabled researchers to quickly develop transgenic lines that improve our understanding of the causes of human blindness and ways to mitigate its effects.
I. Introduction
More is known about photoreceptor cells than any other cell in the vertebrate retina. From early studies in psychophysics (Hecht et al., 1942) and on visual pigments (Wald, 1955), to the work on mechanisms of dark adaptation (Dowling, 1963), phototransduction (Yau, 1994), and inherited retinal disease (Dryja and Li, 1995), rod photoreceptors have been central to the understanding of retinal function and hereditary blindness disorders. Many of the mutations known to cause hereditary retinal degeneration affect rod-specific genes or otherwise interfere with rod function. For reasons still not fully understood, rod degeneration often leads to the secondary death of cone photoreceptors and the loss of color vision and eventually all vision. For decades, insights into the mechanisms of rod photoreceptor degeneration came from studies of naturally occurring mutations and gene knock-outs in nocturnal mammalian models such as rats and mice, whose retinas have ~95% rods. More recently, diseases primarily affecting cone photoreceptors, such as achromatopsia, have prompted investigations into the biochemical and physiological properties of cones. The zebrafish is a diurnal animal with a retina containing large numbers of diverse cone subtypes (Branchek, 1984; Branchek and Bremiller, 1984; Fadool, 2003; Raymond et al., 1993, 1995), which make zebrafish an ideal model to study the pathological mechanisms underlying both rod and cone degeneration, as well as to examine the capacity of the vertebrate retina to regenerate rods and cones.
A. Photoreceptor Anatomy and Biochemistry
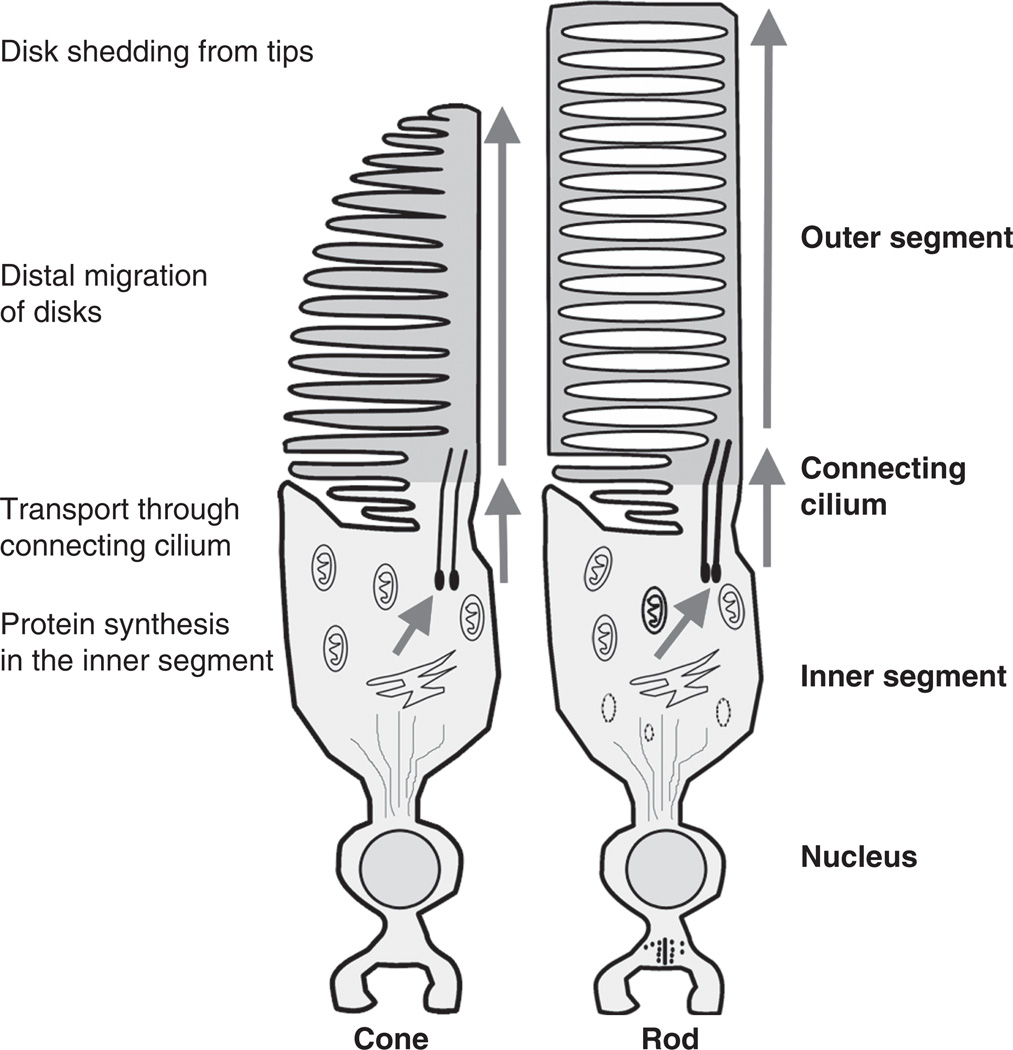
Both rod and cone photoreceptors are highly specialized cells with a unique morphology consisting of an elongated outer segment, connecting cilium, inner segment, cell body, and synaptic terminal (Fig. 1). The shape and morphology of the outer segments (OSs) usually distinguish the rods from the cones and provide the basis for their nomenclature. The OS consists of hundreds of tightly stacked membrane discs that contain the proteins necessary for phototransduction. Protein synthesis occurs in the inner segment, and molecules destined for the OS must be transported apically through the connecting cilium via a process known as intraflagellar transport (IFT) (see Insinna and Besharse, 2008 for a recent review). The inner segment also contains numerous mitochondria needed to provide the energy for the demands of protein synthesis, protein trafficking, and phototransduction. The synaptic terminals of rod photoreceptors, known as spherules, are typically smaller than the cone terminals, known as pedicles. Both spherules and pedicles are filled with synaptic vesicles, contain synaptic ribbons, and are presynaptic to the bipolar and horizontal cells.
Fig. 1.
Anatomy cartoon diagrams of vertebrate cone (left) and rod (right) photoreceptors. Arrows indicate the direction of protein trafficking for most outer segment proteins. The outer segments are shaded pink and cellular processes relevant for outer segment development are outlined on the left.
Zebrafish possess one type of rod photoreceptor and four subtypes of cone photoreceptors. The zebrafish cone subtypes absorb light maximally in the red (570 nm), green (480 nm), blue (415 nm), and ultraviolet (362 nm) regions of the spectrum (Robinson et al., 1993). These cone types are also distinguishable morphologically; short and long single cones contain the ultraviolet (UV)- and blue-absorbing visual pigments, respectively, whereas the red- and green-sensitive visual pigments are found in the principal and accessory members of the double cones (Robinson et al., 1993). The cone photoreceptors are arranged in a highly ordered crystalline mosaic, with rod inner segments projecting through the cone mosaic to surround the UV cones (Fadool, 2003). Additional details about the cone mosaic and cone physiology can be found in two recent review articles (Fadool and Dowling, 2008; Raymond and Barthel, 2004)
B. Photoreceptor Development
In vertebrates, neurogenesis of the retina begins when proliferating neuroblasts exit the cell cycle and begin the process of differentiation. In most species, including zebrafish, cone differentiation occurs before rod differentiation. The expression of the zebrafish rod opsin gene begins at approximately 50 h post-fertilization (hpf) (Schmitt and Dowling, 1996) in a region of precocious neurogenesis in the ventral nasal retina referred to as the ventral patch. The red and blue cone opsin genes are also expressed at about 50 hpf while expression of the UV opsin gene is observed about 5 h later (Schmitt et al., 1999). Similarly, small clusters of rods and the red/green double cones are labeled with the monoclonal antibodies ROS-1 and Zpr-1, respectively, in whole mount at 50 hpf (Raymond et al., 1995). Given that the first cells in the outer nuclear layer (ONL) of the retina to become post-mitotic are seen between 43 and 48 hpf (Hu and Easter, 1999), it is likely that cell cycle exit precedes opsin expression by several hours. Cone differentiation occurs in a sweeping fashion from the ventro-nasal side of the choroid fissure to the dorsonasal retina and then to the dorsotemporal and ventrotemporal side of the choroid fissure (Raymond et al., 1995; Schmitt and Dowling, 1996). On the other hand, rod differentiation initiates in the ventral retina on the nasal side of the choroid fissure but soon crosses directly to the temporal side of the fissure. This ventral patch of rods grows symmetrically across the choroid fissure and increases in density while slowly expanding. Between 50 and 72 hpf, rods fill in the dorsal retina in a seemingly random fashion that does not resemble the wave of differentiation exhibited by all other retinal cell types (Raymond et al., 1995; Schmitt and Dowling, 1996). Despite similarities in timing of early rod and cone differentiation, zebrafish vision is predominantly cone driven for several days. Based upon behavioral studies and electroretinogram (ERG) recordings, rod function can first be demonstrated only between 14 and 21 days post-fertilization (dpf) (Bilotta et al., 2001; Saszik et al., 1999). Spectral sensitivity appears to be cone dominated through 15 dpf, and the rod contribution is not adult-like until close to 1 month of age (Bilotta et al., 2001; Saszik et al., 1999).
A number of intrinsic and extrinsic factors contribute to the specification, differentiation, and maturation of rods, but the story is far from complete. For example, the transcription factors neural retina leucine zipper (Nrl), cone–rod homeobox (crx) gene, neuroD, and tbx2b regulate distinct aspects of rod development (Alvarez-Delfin et al., 2009; Furukawa et al., 1997, 1999; Mears et al., 2001). Extrinsic factors known to promote rod specification and differentiation include sonic hedgehog (Shh) (Shkumatava et al., 2004; Stenkamp and Frey, 2003), taurine (Young and Cepko, 2004), fibroblast growth factor (Patel and McFarlane, 2000), and retinoic acid (Hyatt et al., 1996; Perkins et al., 2002). Given the potentially broad 12-h time window for cell cycle exit and differentiation of photoreceptors (~43–55 hpf), it is unclear if these extrinsic signals contribute equally to the specification and differentiation of rods and cones.
C. Improvements in Transgenic Technology
The techniques to generate transgenic zebrafish have significantly improved in the last 5 years. In the past, linearized plasmid DNA containing the transgene was injected into the yolks of 1–4-cell-stage zebrafish embryos. In a small percentage of cases (typically 5–10%), the DNA randomly integrates into the genome of the germ cells. These “founder fish” produced transgenic progeny. As described in a previous review (Perkins et al., 2004), this approach was used to generate transgenic zebrafish lines that express GFP exclusively in rod photoreceptor cells (Fadool, 2003; Hamaoka et al., 2002; Kennedy et al., 2001; Perkins et al., 2002), one line that expresses GFP in all cone photoreceptors (Kennedy et al., 2007), and one line that selectively marks UV cones (Takechi et al., 2003). Today, transgenesis frequencies have been increased by creating plasmids that contain sequences recognized by a rare-cutting endonuclease, such as I-SceI, or by inserting the transgene into the Tol2 transposon (see Chapter XX). In both cases, the percentage of injected embryos transiently expressing the transgene is elevated severalfold, and the rate of germ line transmission has approached 70% (Balciunas et al., 2006). Construction of Tol2 vectors for transgenesis is now facilitated by the use of site-specific recombination cloning (Gateway®) technology, as part of the “Tol2kit” (Kwan et al., 2007). The Tol2kit consists of various plasmids containing Gateway recombination sequences. A functional Tol2 transposon may be rapidly assembled in a modular fashion by recombining a promoter-containing 5′ element, a 3′-tag, and a Tol2 transposon backbone. With these tools in hand, one can quickly construct Tol2 transposons and generate transgenic lines that express fluorescent markers behind cell-specific or inducible promoters and express proteins fused at the N- or C-terminus with fluorescent markers or protein purification tags, as well as other variations.
II. Transport Mechanisms
Photoreceptor OSs rapidly turn over throughout the lives of photoreceptor cells; thus, efficient protein transport mechanisms provide photoreceptors with a sufficient supply of protein to replenish that lost during OS turnover. It is estimated that rhodopsin constitutes 90% of the total protein in the rod OS and therefore plays important roles in both the physiology and structural integrity of the photoreceptor (Nathans, 1992). As such, investigations into this process have primarily focused on the transport of rhodopsin.
The photoreceptor OS can be considered a specialized sensory cilium that concentrates opsin for light detection. All cilia consist of a microtubule-based axoneme surrounded by the ciliary membrane. Beginning at the basal body, the photoreceptor axoneme extends into a transition zone, known as the connecting cilium, and continues distally into the OS where it terminates near the tip as singlet microtubules (Insinna and Besharse, 2008). Disc membrane assembly occurs at the base of the OS, just beyond the distal end of the transition zone. A process known as intraflagellar transport, or IFT, transports opsin molecules through the connecting cilium where they then incorporate into newly assembled disc membranes (Insinna et al., 2008, 2009a; Marszalek et al., 2000; Pazour et al., 2002). IFT refers to the bidirectional movement of multisubunit IFT particles along the length of the ciliary axoneme. The IFT particle is composed of at least 18 distinct proteins and perhaps dozens of associated protein components and cargo molecules (Hao and Scholey, 2009). Although most OS proteins incorporate into disc membranes and are eventually shed from the distal tips of photoreceptors, IFT particles are believed to migrate along the length of the axoneme in both an anterograde and a retrograde manner. Evidence for this comes from transient transgenesis experiments where constructs containing IFT proteins fused to GFP were expressed using a zebrafish rhodopsin promoter (Insinna et al., 2009b; Kennedy et al., 2001; Luby-Phelps et al., 2008). Rod-specific overexpression of IFT20-GFP, IFT52-GFP, IFT57-GFP, and IFT88-GFP found localization of IFT proteins along the entire length of the axoneme, as well as the basal body and the connecting cilium.
Cilia formation and anterograde IFT movement require two kinesin motor proteins. The heterotrimeric kinesin-II motor, which is composed of the Kif3a, Kif3b kinesin subunits and a kinesin-associated protein (KAP) subunit, was first shown to be required for rod photoreceptors (Marszalek et al., 2000). Studies in Caenorhabditis elegans, however, revealed that a homodimeric kinesin -II motor, OSM-3, was also required for ciliogenesis in some sensory neurons. Insinna and colleagues (2008, 2009a)) found that Kif17, the vertebrate homolog to OSM-3, was also essential for ciliogenesis and OS formation in zebrafish photoreceptors. The authors compared the roles of these different kinesin motors in photoreceptor function by creating new constructs that utilized the promoter for the zebrafish cone transducin alpha subunit (TαC) (Kennedy et al., 2007) to drive dominant-negative forms of either the Xenopus Kif3b (Lin-Jones et al., 2003) or the mouse Kif17 (Chu et al., 2006). In both cases, the motor domain of the kinesin protein was replaced by GFP. The 3.2 kilobase fragment of the TαC promoter drives expression in all cone subtypes, although the first observed expression of a GFP transgene does not occur until ~70 hpf, which is several hours after endogenous opsin expression (Kennedy et al., 2007). Overexpression of the dominant-negative Kif17 (DNKIF17) caused severe ablation of cone OSs, whereas the dominant-negative Kif3b (DNKIF3B) was less damaging to cones. In contrast, opsin mislocalization was observed in cells expressing DNKIF3B but not DNKIF17, suggesting that these two kinesins regulate trafficking of distinct cargoes (Insinna et al., 2009b)).
III. Regulation of Photoreceptor Size
The length of mature photoreceptor OSs remains almost constant through the daily balancing act of disc membrane renewal. Approximately 10% of the OS is shed from the apical tips and this material is replaced at the OS base on a daily basis. Almost nothing is known about the mechanisms that maintain OS at a constant size or the mechanisms that regulate the IFT process, thereby facilitating OS renewal. Recent work has suggested that the FERM protein Mosaic eyes may function to negatively regulate apical renewal of photoreceptors (Hsu et al., 2006). The zebrafish mosaic eyes (moe) gene is essential for proper retinal lamination (Jensen et al., 2001), thereby preventing analysis of photoreceptor morphology. The lamination defects could be rescued, however, through genetic mosaic analysis, which also revealed cell-autonomous functions for Moe. The rod-specific (Tg)XOPS:GFP transgene was placed on the moe−/− background to help visualize transplanted photoreceptors and to provide a means to measure cell volume. In the wild-type host retinas, GFP-expressing moe−/− rods adopted a normal morphology but the cell volume was nearly 50% greater than wild type at 6 dpf. Most of this increased volume appeared to be due to an increase in OS volume. The Moe protein directly interacted with a number of Crumbs proteins and this interaction suggests a potential regulatory mechanism for OS length. In Drosophila, overexpression of crumbs expands the apical domains of ectodermal epithelia (Wodarz et al., 1995) and lengthens the stalk region of photoreceptors (Pellikka et al., 2002). These results suggest a conserved mechanism whereby Crumbs proteins function positively in OS growth and Moe negatively regulates Crumbs function (Hsu et al., 2006).
IV. Photoreceptor Synapse Structure
Genetic screens utilizing the optokinetic response (OKR) assay have successfully identified mutants that affect synaptic transmission between cone photoreceptors and bipolar cells of the inner retina (Brockerhoff et al., 1995; Muto et al., 2005; Van Epps et al., 2004). One such example is the no optokinetic response c (nrc) mutant, which disrupts the synaptojanin 1 protein. Synaptojanin 1 is a polyphosphoinositide phosphatase that regulates clathrin-mediated endocytosis at conventional synapses. In nrc mutant photoreceptor synapses, the ribbons formed normally but would “float” unanchored from the synaptic junction, fewer synaptic vesicles were present within the synapse, and the arciform density, which anchors the ribbon to the plasma membrane, was missing (Allwardt et al., 2001; Van Epps et al., 2004). Interestingly, changes in synaptic architecture could be observed using confocal microscopy of transgenic mutants (Tg(TαC:GFP)nrc). In wild-type cells expressing the TαC:GFP transgene, the cones form invaginating synapses that appear like a “donut” of fluorescence, with a dark center corresponding to the non-fluorescent bipolar cells. The mutant cone terminals were flattened and lacked the dark center, indicating that nrc mutant cones failed to form invaginating synapses with bipolar cells.
V. Regeneration
Persistent neurogenesis and regeneration in the visual system of adult teleost fish have been valuable models of neural development. Zebrafish, like many teleosts, continue to grow throughout their life, and the increase in body mass is matched by an increase in the size of the eye and the area of the retina (Fernald, 1990; Johns, 1982; Johns and Fernald, 1981; Marcus et al., 1999; Otteson et al., 2001). In the adult, neurogenesis occurs from two distinct stem cell populations (Hecht et al., 1942): multipotent neural progenitors at the retinal margin, which can give rise to all retinal cell types except rods, and (Wald, 1955) slowly dividing stem cells within the ONL that serve as rod precursors. At the retinal margin, mitotic cells possess properties of stem cells, maintaining a balance between the self-renewal and the generation of multipotent neuroblasts that differentiate into all classes of neurons and glia. The spatial expression of proneural genes, cell-to-cell signaling molecules, and cellular differentiation markers recapitulates the temporal sequence observed during embryonic development. The identification of neurogenesis at the margin of the hatchling chick retina and the isolation of proliferative cells from the ciliary margin of rodents suggested an evolutionarily conserved system, although further experimental evidence is warranted (Ahmad, 2001; Cicero et al., 2009; Fischer and Reh, 2000; Haruta et al., 2001; Tropepe et al., 2000).
During post-embryonic growth in teleosts, new rods are generated in the central retina from a population of mitotic cells referred to as the rod progenitor lineage. As the animal grows, the retina is gradually stretched within the expanding optic cup, and cell density decreases for all cells except rod photoreceptors. Visual acuity is maintained by increasing the size of the retinal image proportional to that of the increase in eye size. Visual sensitivity, however, is maintained by the generation of new rods in the central retina from a population of mitotic cells referred to as rod progenitor cells (Johns, 1982; Johns and Fernald, 1981). The addition of new rods ensures a constant density across the ONL, thereby preserving scotopic sensitivity (Fernald, 1990; Johns, 1982). Rod progenitors were initially identified as proliferating cells that were distributed across the ONL and served as a source of newly generated rods (Johns, 1982; Johns and Fernald, 1981). These rod precursors proliferate at a low rate and subsequently differentiate into rod photoreceptors. Subsequent studies using multiple injections of tritiated thymidine or exposure to thymidine analogues revealed groups of mitotically active cells arranged in radial arrays spanning the inner nuclear layer (INL) and ONL. In histological sections, these “neurogenic clusters” appeared to migrate along Müller glia. The author proposed that the clusters of proliferating cells were rod progenitors that migrated to the outer retina and became the source of the rod precursors (Johns, 1982).
Perhaps more interesting is that in teleosts, including zebrafish, cell death resulting from mechanical damage, chemical toxicity, phototoxicity, or genetic lesions stimulates an increase in cellular proliferation in the INL and ONL, followed by regeneration of lost cell types, including damaged rod photoreceptors (Braisted et al., 1994; Fausett and Goldman, 2006; Vihtelic and Hyde, 2000; Wu et al., 2001), and recovery of vision (Mensinger and Powers, 1999). Until recently, the cellular and molecular processes underlying regeneration remained largely unknown; however, transgenic analysis of adult neurogenesis in zebrafish has contributed significantly to the understanding of the origin of the cells that maintain the regenerative potential and the genes that regulate regeneration of photoreceptor cells.
Several lines of evidence have identified a subpopulation of Müller glia as the origin of the “neurogenic clusters” that give rise to both the rod progenitor lineage and the multipotent stem cells observed in regenerating fish retinas (Bernardos et al., 2007; Fausett et al., 2008; Fimbel et al., 2007; Morris et al., 2008; Thummel et al., 2008; Yurco and Cameron, 2005). In both the intact juvenile retina and the following acute photoreceptor cell damage, double-labeling experiments demonstrated that these proliferating cells co-label for definitive markers of Müller cells, including glial fibrillary acid protein (GFAP), carbonic anhydrase, and glutamine synthetase. The most convincing evidence of the single origin for both of these processes was provided by Barnados et al. (Bernardos and Raymond, 2006; Bernardos et al., 2007) who used Tg (gfap:GFP)mi2002 transgenic zebrafish, in which regulatory elements of the zebrafish GFAP gene drove expression of cytoplasmic or nuclear-targeted GFP specifically in Müller glia. The authors used the persistence of GFP fluorescence as a lineage tracer to track the fates of cells derived from the Müller glia. Following bromodeoxyuridine (BrdU) labeling to identify mitotically active cells in the uninjured retina, the authors observed small numbers of BrdUþ cells in the INL and ONL, some of which were also GFPþ. Following light damage, the number of mitotically active (i.e., BrdUþ) GFPþ cells increased considerably and could be traced as clusters extending from the INL to the ONL where some co-labeled for the transcription factor Crx and other markers of differentiating photoreceptors. The authors concluded that the co-labeled cells were dedifferentiated Müller glia and their progeny, which migrate from the INL into the ONL to form rod precursors and regenerated photoreceptors, respectively. It has also been demonstrated that constant intense light treatment of dark-adapted albino zebrafish selectively kills rod and cone photoreceptors in the central retina (Qin et al., 2009; Vihtelic and Hyde, 2000) and induces approximately 50% of the Müller glia to co-label for mitotic markers such as proliferating cell nuclear antigen (PCNA) (Thummel et al., 2008). Injection and electroporation of antisense morpholinos complementary to PCNA prior to retinal lesion led to a significant increase in the number of dying cells in the INL and reduced both the number of proliferating cells and the number of Müller glia in the region of the light damage (Thummel et al., 2008). These data suggest that following retinal lesion, asymmetric cell division of Müller glia generates a mitotic progenitor that gives rise to the neurogenic cluster while maintaining a constant population of Müller glia.
As regeneration progresses, the Müller-derived stem cells expressed numerous genes consistent with a photoreceptor developmental program. Following retinal damage, the Müller glial-derived mitotic cells initiate expression of retinal stem/ progenitor cell markers, including ascl1a, pax6, rx1, neurogenin1, and chx10 (Fausett and Goldman, 2006; Raymond et al., 2006; Thummel et al., 2010). As the retinal progenitors migrate from the INL to the ONL, Pax6 expression is lost and other transcription factors are expressed, most notably the basic helix-loop-helix gene neurod, and the cone–rod homeobox gene crx. Similarly, cell signaling molecules of the Notch–Delta pathway and its downstream effectors are also expressed (Raymond et al., 2006). These data suggest that progenitor cells produced in the INL in response to acute injury pass through a series of intervening, less-committed states prior to adopting a photoreceptor cell fate.
To uncover genes specifically expressed by the Müller glial-derived mitotic progenitors, the gene expression profiles were compared with those of GFPþ cells isolated from intact and light-lesioned Tg(gfap:GFP)mi2002 zebrafish retinas (Qin et al., 2009). Using fluorescence-activated cell sorting, the GFPþ cells were isolated from control retinas and retinas up to 36 h post light damage. Similar to other studies of changes in gene expression following acute damage, gene networks associated with increased proliferation, stress response, and neurogenesis were activated. The regeneration of the retina shares features common to other regenerating tissues including the heart and fin. Two genes required for fin and heart regeneration in zebrafish—hspd1, which encodes heat shock protein 60 and mps1, a protein kinase involved in mitotic checkpoint regulation—are up-regulated in injury-activated Müller glia (Qin et al., 2009). Co-labeling confirmed that both genes were expressed in the mitotically activated Müller glia and their progeny. Quite fortuitously, mutant alleles exist in zebrafish for both of these genes. The mutant no blastema (nbl) is a temperature-sensitive null allele of hspd1 that disrupts chaperone activity (Makino et al., 2005). nightcap (ncp) has a missense substitution in the conserved kinase domain of mps1 and also exhibits a temperature-sensitive phenotype (Poss et al., 2002). By rearing fish at the non-permissive temperature, the authors demonstrated that both were required at different stages of cone photoreceptor regeneration following light damage.
Whereas current work in numerous species has focused on the Müller cells and the INL stem cells, understanding the regulation of rod progenitor cell activity may offer an alternative avenue to investigate the fundamental processes of photoreceptor replacement. Two previous limitations of such studies have been that rod precursors are relatively few in number and no definitive marker specific for the rod progenitors is available. The XOPS-mCFP transgenic line experiences selective degeneration of the rod photoreceptor cells due to the toxic effects of high-level expression of a rod-targeted fluorescent reporter gene (Morris and Fadool, 2005). This rod degeneration resulted in a loss of rod-mediated electrophysiological responses, but did not cause any secondary cone pathology. It was, however, accompanied by a significant increase in rod progenitor mitotic activity (Morris and Fadool, 2005). Similar to cone regeneration, the transcription factors NeuroD and Crx were upregulated in cells in the ONL. But, in contrast, expression of nr2e3—a rod determination gene found exclusively in post-mitotic differentiating rod photoreceptors—was also upregulated, whereas expression of pax6 was not observed (Morris and Fadool, 2005). Immunolabeling of retinal cryosections with anti-BrdU showed no significant increase in INL BrdUþ cells or PCNA expression. The expression of genes with demonstrated roles in photoreceptor development, such as neurod, crx, and nr2e3, combined with the lack of pax6 expression, suggests an early commitment of the mitotic rod precursors to the rod cell fate (Morris and Fadool, 2005). Supporting this conclusion, knock-down of both Pax6a and Pax6b resulted in increased numbers of rod precursors in the ONL without affecting rod regeneration (Thummel et al., 2010). Therefore, it appears that the rod progenitors in the ONL maintain the capacity to respond to rod photoreceptor degeneration without relying on increased activity of progenitor cells in the INL or the Müller glia. By contrast, cone-specific cell death resulting from mutation of the pde6c gene stimulates Müller glial proliferation in the absence of rod cell death (Morris et al., 2008) which we inferred that the retina responds to rod and cone cell death differently.
Others have demonstrated mitosis of Müller glia following a specific form of rod death. Regeneration following acute and specific loss of all mature rod photoreceptors was tested using the bacterial nitroreductase (NTR)/metronidazole cell ablation system (Montgomery et al., 2010). The authors describe two independent transgenic lines of zebrafish that express an NTR-EGFP fusion protein from the zebrafish rod opsin promoter. Similar lines have also been generated to induce the specific loss of bipolar cells in the retina (Montgomery et al., 2010). In the former studies, uniform expression of NTR-EGFP in all rod photoreceptors was observed in the Tg(zop:nfsB-EGFP)nt19 line, whereas a subset of rods express NTR-EGFP in Tg(zop:nfsB-EGFP)nt20 line. Treatment with metronidazole resulted in the death of all rods expressing the NTREGFP fusion protein. In contrast to what we observed in the Tg(xops:mCFP) transgenic line, metronidazole treatment of the Tg(zop:nfsB-EGFP)nt19 fish increased Müller glial cell proliferation. However, the authors reported that only increased rod precursor cell proliferation was observed following treatment of the Tg(zop:nfsB-EGFP)nt20 fish. Thus, acute loss of the majority of rod photoreceptors was sufficient to induce a Müller glial regeneration response, suggesting that the level of cell death is an important factor regulating increased proliferation of Muller glial versus rod progenitors.
These studies highlight the significant gains made possible by combining advances in many areas of zebrafish genetics such as the development of novel transgenic lines, application of cell sorting, gene profiling arrays, and cell ablation technologies. The ever-increasing utility of transgenic tools to investigate photoreceptor biology in zebrafish has mirrored many of the advances in other organ systems. The continuation of these studies should allow a more detailed characterization of the intrinsic properties of retinal stem cells, identification of the environmental signals that direct regeneration of the photoreceptor cells, and development of methods to manipulate cells to direct photoreceptor cell replacement.
VI. Future Directions
Transgenic zebrafish are powerful tools to analyze photoreceptor morphology, cell biology, and regeneration. To date, studies have utilized transgenic technology to express intrinsic proteins behind very strong promoters (e.g., Xenopus opsin or cone transducin promoters). While deleterious effects of overexpressing some proteins have not yet been reported, it is likely that overexpression of some proteins cannot be tolerated by rods or cones. Future efforts to identify and characterize weakly and moderately expressing promoters will enhance the toolkit available to zebrafish researchers and reduce the chances of overexpression toxicity.
The role of extrinsic factors on photoreceptor development and differentiation has largely relied on loss of function strategies (e.g., mutagenesis and morpholino approaches) and overexpression following mRNA injection. In both cases, analysis on photoreceptors is often limited because many signaling molecules have essential roles much earlier in development and the embryos may not survive to later time points, or photoreceptor phenotypes may be reflect non-specific effects from general developmental abnormalities. Temporally regulated expression of transgenes is one potential method to bypass these limitations. The promoter of the heat shock protein, hsp70, allows transgene expression to be limited to those times when the animals are exposed to a brief heat shock. This temporal control of transgene expression can be used to drive expression of signaling molecules or dominant-negative forms of growth factor receptors during windows critical for photoreceptor development but well after critical developmental events, such as gastrulation or optic cup formation. These approaches will significantly enhance our understanding of how some molecules contribute to both photoreceptor differentiation and regeneration.
VII. Conclusions
The study of photoreceptor cell structure and development has benefited from the use of transgenic zebrafish expressing cell-specific GFP reporter genes. At the moment there are several rod- and cone-specific GFP lines, and one cone subtype-specific line, namely that for UV opsin-expressing cones. Transgenic technology will greatly facilitate future studies of neuronal structure and function in zebrafish. Dozens of different transgenic zebrafish lines currently exist (Udvadia and Linney, 2003) and more specialized lines will no doubt appear in the future. The classic studies of Ramón y Cajal used Golgi staining to provide tremendous insights into the structure of retinal neurons in fixed tissue (Ramon and Cajal, 1911). With the increasing power of fluorescent imaging technology, questions about the development and function of neurons in vivo and in real time are being addressed. With the appropriate promoters, it should be possible to generate individual transgenic lines that express reporter genes within all subsets of the major classes of retinal cells. These lines could be used for rapid identification of specific cell types prior to electrophysiological recordings. Additionally, time-lapse imaging of individual cells or whole layers of cells could be used to study the timing and control of synaptogenesis in vivo, as seen from the report by Kay et al. (2004). Certainly the combination of these techniques with the wide assortment of zebrafish mutants will facilitate our understanding of neural development and function.
References
- Ahmad I. Invest. Ophthalmol. Vis. Sci. 2001;42:2743–2748. [PubMed] [Google Scholar]
- Allwardt BA, Lall AB, Brockerhoff SE, Dowling JE. J. Neurosci. 2001;21:2330–2342. doi: 10.1523/JNEUROSCI.21-07-02330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Delfin K, Morris AC, Snelson CD, Gamse JT, Gupta T, Marlow FL, Mullins MC, Burgess HA, Granato M, Fadool JM. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2023–2028. doi: 10.1073/pnas.0809439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. J. Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. Gene Expr. Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bilotta J, Saszik S, Sutherland SE. Dev. Dyn. 2001;222:564–570. doi: 10.1002/dvdy.1188. [DOI] [PubMed] [Google Scholar]
- Braisted JE, Essman TF, Raymond PA. Development. 1994;120:2409–2419. doi: 10.1242/dev.120.9.2409. [DOI] [PubMed] [Google Scholar]
- Branchek T. J. Comp. Neurol. 1984;224:116–122. doi: 10.1002/cne.902240110. [DOI] [PubMed] [Google Scholar]
- Branchek T, Bremiller R. J. Comp. Neurol. 1984;224:107–115. doi: 10.1002/cne.902240109. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, Dowling JE. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10545–10549. doi: 10.1073/pnas.92.23.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PJ, Rivera JF, Arnold DB. J. Biol. Chem. 2006;281:365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- Cicero SA, Johnson D, Reyntjens S, Frase S, Connell S, Chow LM, Baker SJ, Sorrentino BP, Dyer MA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JE. J. Gen. Physiol. 1963;46:1287–1301. doi: 10.1085/jgp.46.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Li T. Hum. Mol. Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- Fadool JM. Dev. Biol. 2003;258:277–290. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Dowling JE. Prog. Retin. Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. J. Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D. J. Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernald RD. J. Exp. Zool. Suppl. 1990;5:167–180. doi: 10.1002/jez.1402560521. [DOI] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. J. Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Dev. Biol. 2000;220:197–210. doi: 10.1006/dbio.2000.9640. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Nat. Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S. Genesis. 2002;34:215–220. doi: 10.1002/gene.10155. [DOI] [PubMed] [Google Scholar]
- Hao L, Scholey JM. J. Cell. Sci. 2009;122:889–892. doi: 10.1242/jcs.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Kosaka M, Kanegae Y, Saito I, Inoue T, Kageyama R, Nishida A, Honda Y, Takahashi M. Nat. Neurosci. 2001;4:1163–1164. doi: 10.1038/nn762. [DOI] [PubMed] [Google Scholar]
- Hecht S, Schlaer S, Pirenne MH. J. Optic. Soc. Amer. 1942;38:196–208. [Google Scholar]
- Hsu YC, Willoughby JJ, Christensen AK, Jensen AM. Development. 2006;133:4849–4859. doi: 10.1242/dev.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Easter SS. Dev. Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- Hyatt GA, Schmitt EA, Fadool JM, Dowling JE. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13298–13303. doi: 10.1073/pnas.93.23.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Besharse JC. Dev. Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse JC. Dev. Dyn. 2009a doi: 10.1002/dvdy.21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Luby-Phelps K, Link BA, Besharse JC. Methods Cell Biol. 2009b;93:219–234. doi: 10.1016/S0091-679X(08)93012-0. [DOI] [PubMed] [Google Scholar]
- Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. Dev. Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AM, Walker C, Westerfield M. Development. 2001;128:95–105. doi: 10.1242/dev.128.1.95. [DOI] [PubMed] [Google Scholar]
- Johns PR. J. Neurosci. 1982;2:178–198. doi: 10.1523/JNEUROSCI.02-02-00178.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns PR, Fernald RD. Nature. 1981;293:141–142. doi: 10.1038/293141a0. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Alvarez Y, Brockerhoff SE, Stearns GW, Sapetto-Rebow B, Taylor MR, Hurley JB. Invest. Ophthalmol. Vis. Sci. 2007;48:522–529. doi: 10.1167/iovs.06-0975. [DOI] [PubMed] [Google Scholar]
- Kennedy BN, Vihtelic TS, Checkley L, Vaughan KT, Hyde DR. J. Biol. Chem. 2001;276:14037–14043. doi: 10.1074/jbc.M010490200. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lin-Jones J, Parker E, Wu M, Knox BE, Burnside B. Invest. Ophthalmol. Vis. Sci. 2003;44:3614–3621. doi: 10.1167/iovs.03-0164. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K, Fogerty J, Baker SA, Pazour GJ, Besharse JC. Vision Res. 2008;48:413–423. doi: 10.1016/j.visres.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Whitehead GG, Lien CL, Kim S, Jhawar P, Kono A, Kawata Y, Keating MT. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14599–14604. doi: 10.1073/pnas.0507408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Delaney CL, Easter SS., Jr Vis. Neurosci. 1999;16:417–424. doi: 10.1017/s095252389916303x. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Liu X, Roberts EA, Chui D, Marth JD, Williams DS, Goldstein LS. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nat. Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Mensinger AF, Powers MK. Vis. Neurosci. 1999;16:241–251. doi: 10.1017/s0952523899162059. [DOI] [PubMed] [Google Scholar]
- Montgomery JE, Parsons MJ, Hyde DR. J. Comp. Neurol. 2010;518:800–814. doi: 10.1002/cne.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Fadool JM. Physiol. Behav. 2005;86:306–313. doi: 10.1016/j.physbeh.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AC, Scholz TL, Brockerhoff SE, Fadool JM. Dev. Neurobiol. 2008;68:605–619. doi: 10.1002/dneu.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, Gahtan E, Xiao T, Nevin LM, Gosse NJ, Staub W, Finger-Baier K, et al. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J. Biochemistry. 1992;31:4923–4931. doi: 10.1021/bi00136a001. [DOI] [PubMed] [Google Scholar]
- Otteson DC, D’Costa AR, Hitchcock PF. Dev. Biol. 2001;232:62–76. doi: 10.1006/dbio.2001.0163. [DOI] [PubMed] [Google Scholar]
- Patel A, McFarlane S. Dev. Biol. 2000;222:170–180. doi: 10.1006/dbio.2000.9695. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. J. Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- Perkins BD, Fadool JM, Dowling JE. Methods Cell Biol. 2004;76:315–331. doi: 10.1016/s0091-679x(04)76015-x. [DOI] [PubMed] [Google Scholar]
- Perkins BD, Kainz PM, O’Malley DM, Dowling JE. Vis. Neurosci. 2002;19:257–264. doi: 10.1017/s0952523802192030. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Qin Z, Barthel LK, Raymond PA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon Y, Cajal S. Histologie du Systeme Nerveax de l’Homme et des Vertebres. 1911 [Google Scholar]
- Raymond PA, Barthel LK. Int. J. Dev. Biol. 2004;48:935–945. doi: 10.1387/ijdb.041873pr. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. BMC Dev. Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Curran GA. J. Comp. Neurol. 1995;359:537–550. doi: 10.1002/cne.903590403. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Rounsifer ME, Sullivan SA, Knight JK. Neuron. 1993;10:1161–1174. doi: 10.1016/0896-6273(93)90064-x. [DOI] [PubMed] [Google Scholar]
- Robinson J, Schmitt EA, Harosi FI, Reece RJ, Dowling JE. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6009–6012. doi: 10.1073/pnas.90.13.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saszik S, Bilotta J, Givin CM. Vis. Neurosci. 1999;16:881–888. doi: 10.1017/s0952523899165076. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. J. Comp. Neurol. 1996;371:222–234. doi: 10.1002/(SICI)1096-9861(19960722)371:2<222::AID-CNE3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Hyatt GA, Dowling JE. Vis. Neurosci. 1999;16:601–605. doi: 10.1017/s0952523899163181. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Fischer S, Muller F, Strahle U, Neumann CJ. Development. 2004 doi: 10.1242/dev.01247. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Frey RA. Dev. Biol. 2003;258:349–363. doi: 10.1016/s0012-1606(03)00121-0. [DOI] [PubMed] [Google Scholar]
- Takechi M, Hamaoka T, Kawamura S. FEBS Lett. 2003;553:90–94. doi: 10.1016/s0014-5793(03)00977-3. [DOI] [PubMed] [Google Scholar]
- Thummel R, Enright JM, Kassen SC, Montgomery JE, Bailey TJ, Hyde DR. Exp. Eye. Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR. Exp. Eye. Res. 2008;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, van der Kooy D. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Linney E. Dev. Biol. 2003;256:1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Van Epps HA, Hayashi M, Lucast L, Stearns GW, Hurley JB, De Camilli P, Brockerhoff SE. J. Neurosci. 2004;24:8641–8650. doi: 10.1523/JNEUROSCI.2892-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. J. Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Wald G. Am. J. Ophthalmol. 1955;40:18–41. doi: 10.1016/0002-9394(55)91835-3. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Wu DM, Schneiderman T, Burgett J, Gokhale P, Barthel L, Raymond PA. Invest Ophthalmol. Vis. Sci. 2001;42:2115–2124. [PubMed] [Google Scholar]
- Yau KW. Invest. Ophthalmol. Vis. Sci. 1994;35:9–32. [PubMed] [Google Scholar]
- Young TL, Cepko CL. Neuron. 2004;41:867–879. doi: 10.1016/s0896-6273(04)00141-2. [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA. Vision Res. 2005;45:991–1002. doi: 10.1016/j.visres.2004.10.022. [DOI] [PubMed] [Google Scholar]