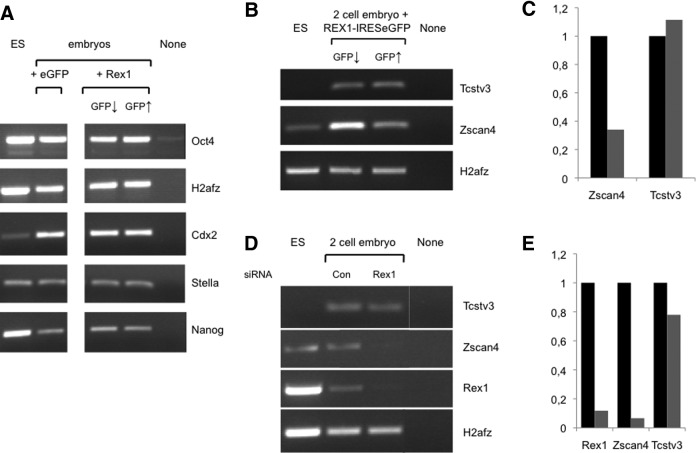
FIG. 7.
Gene expression in Rex1 gain- and loss-of-function mouse embryos. (A) One-cell embryos were microinjected (day 1) with plasmids that direct expression of REX1-IRESeGFP or eGFP as a control, and embryos were separated in (GFP↑) or (GFP↓) groups as explained in the legend to Fig. 6A. RNA levels were analyzed by semiquantitative RT-PCR in embryos taken on day 5. RT-cDNA from equivalent amounts of embryos was amplified with primers specific for the genes indicated; H2afz was used as a control for input. The figure shows an ethidium bromide-stained gel scan from a representative experiment. RNA extracted from E14T ES cells was processed alongside as a positive control. The following templates were used for amplification: E14T ES cells (ES), embryos overexpressing eGFP as a control (eGFP), embryos with low or neglectable Rex1 overexpression levels (GFP↓), embryos with high Rex1 overexpression (GFP↑), or Milli-Q water as a negative control (none). (B–E) Deregulation of Zscan4. (B) Embryos were injected on day 1, separated in (GFP↑) and (GFP↓) groups on day 2, and RNA levels were analyzed by semiquantitative RT-PCR in embryos taken on day 2 as described in the legend to Fig. 7A. The figure shows an ethidium bromide-stained gel from 1 representative experiment out of 2 performed. None stands for no cDNA input. (C) Quantification of data shown in (B). Expression levels are represented relative to H2afz levels (gray and black bars for GFP↑and GFP↓, respectively). (B) Embryos were injected with siRNAs and analyzed as described in the legend to Fig. 4B (si). (E) Quantification of data shown in (D). Expression levels are represented relative to H2afz levels (gray and black bars for Rex1 and Control, respectively).