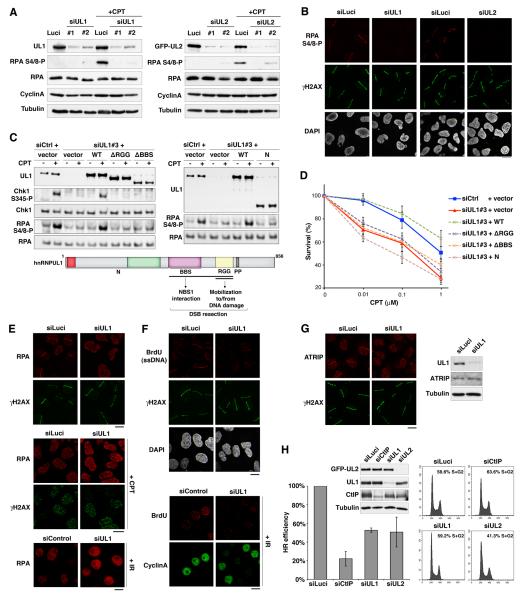
Figure 4. hnRNPUL depletion impairs DNA end resection and HR.
(A) Western-Blot analysis of RPA phosphorylation (RPA S4/8-P) 90 min after camptothecin treatment (+CPT) on total extracts from U2OS cells treated with the indicated siRNAs (Luci: control). U2OS cells stably expressing GFP-hnRNPUL2 were used to assess hnRNPUL2 depletion (right panel). Cyclin A levels monitor S/G2 cells and Tubulin is used as loading control.
(B) Immunodetection of phosphorylated RPA (RPA S4/8-P) at laser micro-irradiation sites in U2OS cells treated with the indicated siRNAs.
(C) Western blotting of total extracts from HeLa cells transfected with the indicated siRNAs and plasmids (vector alone or hnRNPUL1 encoding plasmids) after exposure to 1 μmM CPT for 1 h. siCtrl : control ; siUL1#3 targets hnRNPUL1 3’UTR; hnRNPUL1 mutants refer to the scheme below.
(D) Clonogenic survival of HeLa cells transfected as in C in response to CPT. Error bars: s.e.m. from 3 separate experiments.
(E-F) hnRNPUL1 depletion in U2OS (top panels) and HeLa cells (bottom panel) prevents laser-, CPT- and IR-induced accumulation of RPA and ssDNA formation (BrdU) at DNA damage sites (γH2AX), which is normally observed in S/G2 cells (Cyclin A-positive).
(G) Immuno-detection of ATRIP at laser-damage sites (left) and by western-blotting of total extracts (right) from U2OS cells treated with indicated siRNAs. Cells were analyzed 1 h after micro-irradiation in all cases. Scale bars, 10 μm.
(H) HR-mediated gene conversion assay. HR efficiencies are normalized to siLuci. Error bars: s.d. from 3 independent experiments. CtIP depletion is used as a positive control. siRNA efficiencies are shown on the western-blot panel. Proportions of cells in S/G2 on the corresponding FACS profiles were estimated by the Dean/Jett/Fox model with FlowJo software. See also Figures S4 and S5.