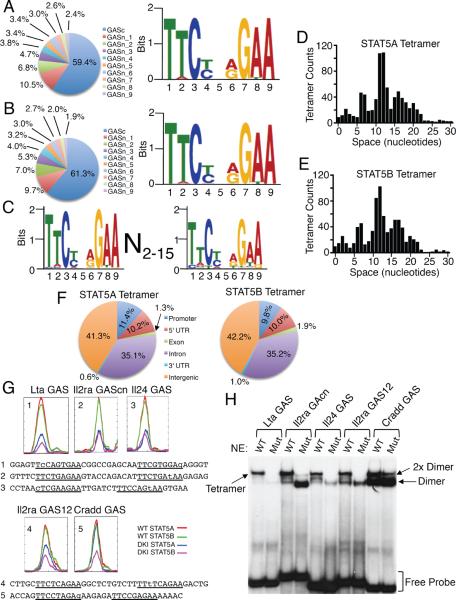
Figure 4. Distribution of STAT5 binding sites and STAT5 dimer and tetramer motifs.
(A and B) Consensus dimer motifs for STAT5A (A) and STAT5B (B). The pie graphs show the top 10 binding motifs for STAT5A (A) and STAT5B (B), as detailed in Supplemental Figure S4D and S4E. (C) Consensus tetramer motif for STAT5A and STAT5B. (D and E) Spacing distribution between tandemly-linked GAS or GAS-like motifs for STAT5A (D) and STAT5B (E). (F) Genome-wide distributions of STAT5A (top) and STAT5B (bottom) tetramer binding sites. 5' UTR, 3' UTR, promoter, and intergenic regions were defined according to Ref Seq data base (February 2006 assembly), and promoters defined as 5 kb upstream of TSS. (G) STAT5 binding motifs for Lta GAS, Il2ra GAScn, Il24 GAS, Il2ra GAS12, and Cradd GAS identified by ChIP-Seq analysis. Color-coded lines indicate chromatin from WT (red and green) or DKI (blue and magenta) T cells immunoprecipitated with anti-STAT5A (red and blue) and STAT5B (green and magenta). Below the graphs are the sequences of the probes for Lta GAS (1), Il2ra GAScn (2) Il24 GAS (3), Il2ra GAS12 (4), and Cradd GAS (5); the GAS and GAS-like motifs are underlined and mismatches are in lower case. (H) EMSA using WT or tetramer-defective mutant STAT5B proteins and GAS motifs from Lta, Il24, Il2ra, and Cradd, based on ChIP-Seq analysis. Tetramer, 2× dimer, dimers, and free probes are indicated based on mobility and whether or not a given probe binds to mutant STAT5B protein. The experiment in panel H was performed 3 times.