Abstract
Plasmodium vivax infections remain a major source of malaria-related morbidity and mortality. Early and accurate diagnosis is an integral component of effective malaria control programs. Conventional molecular diagnostic methods provide accurate results but are often resource-intensive, expensive, have a long turnaround time and are beyond the capacity of most malaria-endemic countries. Our laboratory has recently developed a new platform called RealAmp, which combines loop-mediated isothermal amplification (LAMP) with a portable tube scanner real-time isothermal instrument for the rapid detection of malaria parasites. Here we describe new primers for the detection of P. vivax using the RealAmp method. Three pairs of amplification primers required for this method were derived from a conserved DNA sequence unique to the P. vivax genome. The amplification was carried out at 64°C using SYBR Green or SYTO-9 intercalating dyes for 90 minutes with the tube scanner set to collect fluorescence signals at 1-minute intervals. Clinical samples of P. vivax and other human-infecting malaria parasite species were used to determine the sensitivity and specificity of the primers by comparing with an 18S ribosomal RNA-based nested PCR as the gold standard. The new set of primers consistently detected laboratory-maintained isolates of P. vivax from different parts of the world. The primers detected P. vivax in the clinical samples with 94.59% sensitivity (95% CI: 87.48–98.26%) and 100% specificity (95% CI: 90.40–100%) compared to the gold standard nested-PCR method. The new primers also proved to be more sensitive than the published species-specific primers specifically developed for the LAMP method in detecting P. vivax.
Introduction
Malaria remains one of the top five causes of death in low income countries and is a major source of morbidity worldwide [1]. In 2010, the World Health Organization (WHO) estimated that 216 million clinical cases and 655,000 deaths were due to malaria infections [2]. Geographically, Plasmodium vivax is the most widely distributed human-infecting species, with up to 2.6 billion people at risk for infection [3]. Historically, P. vivax infections have been viewed as “benign” and self-limiting [4], [5]. However, recent evidence suggests that P. vivax infection causes significant morbidity including severe illness leading to mortality [5]–[7]. P. vivax can also co-occur with other Plasmodium species causing further complications for accurate diagnosis and treatment [8].
Early and accurate diagnosis of malaria is essential for case management and for malaria control programs [9]. Current diagnostic methods include microscopy, immunochromatographic rapid diagnostic tests (RDTs) that target parasite proteins, and polymerase chain reaction (PCR) -based molecular tests. Each method has advantages and limitations. Microscopy is considered the gold standard of malaria diagnosis in endemic countries. Correctly employed, it can identify different species and quantitate infection levels. While inexpensive, microscopy is time-consuming, labor-intensive, and it requires well-trained personnel along with appropriate infrastructure [10]. RDTs are easy to use, provide quick results, and are useful alternatives when there is no access to microscopic diagnosis [11]. One major limitation is that most RDTs cannot readily distinguish P. vivax from other species. Molecular diagnostic techniques based on DNA amplification such as PCR-based assays (standard, nested, and real-time) are highly sensitive and retain the ability to accurately differentiate between species [10]. However, since these methods require technical expertise, a well-equipped laboratory, and have long turnaround times, they are beyond the capacity of most malaria-endemic countries, making them difficult to implement in simple clinical laboratories or field settings.
The loop-mediated isothermal amplification (LAMP) method shows great potential for field use as it is simple and field-amenable [12]. DNA is amplified under isothermal conditions, removing the need for sophisticated and expensive thermal cyclers. LAMP employs autocycling strand displacement DNA synthesis using Bacillus stearothermophilus (Bst) DNA polymerase and a set of four primers that bind to unique sites on the target sequence making them highly specific [12]. The use of two loop primers shortens the time to product formation, thereby reducing overall reaction time to between 30 minutes and an hour [13]. LAMP has been utilized to detect malaria parasites mainly using the conventional 18S ribosomal RNA (18S rRNA) gene as the target sequence [14]–[18].
Recently, our laboratory developed a simple portable device that integrates the isothermal amplification technique (LAMP) with a fluorescent detection unit. This new platform, capable of real-time data collection, is referred to as RealAmp. It was used to successfully detect malaria parasites using published Plasmodium genus-specific primers [19]. In this study, we have successfully developed and tested novel P. vivax-specific LAMP primers on the RealAmp platform. These primers target dispersed genomic repeats in the P. vivax genome. Repeats were identified using recently developed diagnostic target mining methods [20].
Methods
Ethics Statement
Clinical samples were retrospectively used in this investigation that were obtained from a molecular surveillance study conducted in Venezuela. This study was originally approved by the institutional ethical review board of Instituto de Altos Estudios Dr. Arnoldo Gabaldon, Maracay, Aragua State, Venezuela. A written consent was obtained from the participants. Some of the clinical samples were obtained from a malaria reference laboratory in the Centers for Disease Control and Prevention (CDC).
Plasmodium clinical samples and laboratory isolates
We used laboratory-adapted malaria parasites and clinical samples obtained from two different sources to test the novel primers. The P. falciparum (3D7) isolate, and other human malaria parasites used in this study such as P. vivax, P. malariae, P. ovale and P. knowlesi were available in the CDC malaria branch collection. Thirteen geographically diverse P. vivax strains (Miami, El Salvador, Honduras, Mauritania, Indonesia, India, South Vietnam, North Korea, Thailand, New Guinea, Panama, Chesson, and Pakchong) were included. Due to genetic similarity between P. vivax and some simian malaria parasites, we also tested eight simian malaria parasites, P. simiovale, P. inui, P. coatneyi, P. hylobati, P. fragile, P. gonderi, P. fieldi, and P. cynomolgi. To determine the limits of detection, we used two fold-serial dilutions of two P. vivax strains (Brazil and India) starting from 2000 parasites/µl to 0.98 parasites/µl. Sixty eight P. vivax clinical samples were obtained from a previous surveillance study conducted in Venezuela. In addition, DNA from 52 clinical samples from our reference diagnostic laboratory, previously diagnosed using a nested-PCR method (10 malaria-negative samples, 14 P. falciparum, 9 P. vivax, 1 P. malariae, 12 P. ovale, 2 P. falciparum/P. malariae, 1 P.vivax/P.ovale, 2 P. falciparum/P. ovale mixed infections and 1 P. knowlesi), were included to test for specificity and sensitivity.
DNA extraction
DNA from blood samples was extracted using the QIAamp DNA minikit (Qiagen, Valencia CA) according to the manufacturer's recommendations. All extracted samples were aliquoted and stored at −20°C.
Nested PCR
Nested PCR designed to amplify 18S ribosomal RNA gene sequences was performed as previously described [19], [21]. Briefly, 20 µl amplification reactions containing 1X buffer, 2.5 mM MgCl2, 200 µM dNTPs, 200 nM primers, and 1.25 units of Taq Polymerase (New England Biolabs, Ipswich, MA) were performed on all samples. Nested-PCR products were analyzed via gel electrophoresis (2% gel) in order to visualize and size the bands.
Data harvesting
Demas et al. applied highly specific criteria to screen and identify unique target sequences for P. vivax and P. falciparum through genome sequence data mining [20]. We used two previously identified P. vivax sequences (Pvr47 and Pvr64) to design novel P. vivax-specific LAMP primers. Several potential P. vivax primers were designed using Eiken's PrimerExplorer software v3 (http://loopamp.eiken.co.jp/e/lamp/primer.html). Five primer sets were tested on the RealAmp platform at temperatures ranging from 60°C to 64°C. Primer sets with the shortest amplification time that correctly amplified P. vivax with no cross-reactivity with other human-infecting or primate Plasmodium species were selected for further analysis.
RealAmp Method
The RealAmp method used for this study has been described in detail [19]. Briefly, the LAMP assays were performed in a 12.5 µL total volume containing a 2X in-house buffer (40 mM Tris-HCl pH 8.8, 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)SO4, 0.2% Tween-20, 0.8 M Betaine, 2.8 mM of dNTPs each), 0.25 µL of a 1∶100 dilution SYBR green, 8 units of Bst DNA polymerase (New England Biolabs, Ipswich, MA), and 2 µl of template DNA. Amplifications were carried out at 64°C for 90 minutes using the ESE-Quant Tube scanner (Qiagen, USA) collecting fluorescence signals at 1-minute intervals. All samples were run at least twice using the r64 primer set as well as published P. vivax LAMP primers [15]. If the two runs were discordant, the sample was run a third time and a criterion of two concordant results out of three runs was used to make the final call. We used a cutoff of 60 minutes and any sample that amplified after this cutoff point was considered to be negative. Since SYBR Green has known potential inhibitory effects on DNA amplification [22], [23], all samples were also tested using 1∶400 dilution of SYTO-9 fluorescent dye.
Statistical Analysis
A total of 120 and 118 samples were included in the sensitivity and specificity analyses of the novel Pvr64 and published P. vivax primers [15], respectively. Nested PCR as described by Singh et al. provided a gold standard [21]. Specificity and sensitivity were calculated as follows:
Sensitivity = true positives/(true positives + false negatives) X 100.
Specificity = true negatives/(true negatives + false positives) X 100.
95% confidence intervals (95% CI) were also calculated.
Results
Novel candidates for primer design
The Pvr47 sequence is described in Demas et al [20]. Pvr64 is a highly conserved repeat family (>98% identity) present in six copies in the P. vivax genome. Five copies are on small contigs that were not assembled into chromosome scaffolds because of their repetitive nature [24]. The remaining copy is on chromosome 9, ∼500 kb from the end of the assembly. Three of the copies are located inside the annotated proteins PVX_092555, PVX_246295, and PVX_238290. PVX_092555 is Plasmodium-specific and has orthologs in the human-infecting species P. falciparum and P. knowlesi, as well as the rodent-infecting species P. berghei, P. chabaudi, and P. yoelli. PlasmoDB does not contain orthologs for PVX_246295 or PVX_238290 [25], but both appear to be truncated paralogs of PVX_092555 based on their nearly identical sequences.
The Pvr64 family is contained within the WD40 domain of P. vivax proteins and most of its Plasmodium orthologs. WD40 domains are found in many proteins implicated in a variety of functions across eukaryotes, including signal transduction, transcriptional regulation, and cell-cycle control. Annotations for most PVX_092555 orthologs contain little functional information, other than to note that they are conserved across the genus Plasmodium. One P. yoelli ortholog is thought to be a transcription factor-associated protein, while another P. yoelli ortholog (which has very low sequence similarity to PVX_092555 and does not contain a WD40 domain) is annotated as an erythrocyte membrane protein.
The Pvr64 family has a high degree of identity with its Plasmodium orthologs (nucleotide identity ranges from 68% in P. falciparum to 86% in P. knowlesi). Because of this high similarity, the Pvr64 segment used to design the novel primers was aligned with the P. knowlesi and P. falciparum sequences and the locations of the six LAMP primers are indicated. Several nucleotide differences were observed within the region used for the six primers (Figure S1).
P. vivax primers
Five primer sets were designed from the two selected target candidates (Pvr64 and Pvr47). Each primer set was first tested with all known human infecting Plasmodium species. If it was able to correctly amplify P. vivax and no other species, then we evaluated it on geographically diverse strains. Based on these criteria, only one primer set based on the Pvr64 target (r64) was selected for further validation (Table 1). The other four primers did not give consistent amplification of the P. vivax DNA and resulted in either very late amplification or negative results, implying that these were not good enough.
Table 1. P. vivax r64 RealAmp primer set sequence (5′→3′).
| F3 | TCT GTT GGT GGA GTA GAT CC |
| B3 | CCT ACG TTT TGG TGA ATC G |
| FIP | ATA TGG TCT CTC GAC ACG GC – CAA ATT GCC ATC ATC TTC AC |
| BIP | TGT GCC CAC CCA CAT ACT TA – GGG AAA TGT TAA TGG GGA TGT |
| LF | AGG CTA CTT CTT TTG CTC C |
| LB | ACT TAC AGT GCT GTA GAG A |
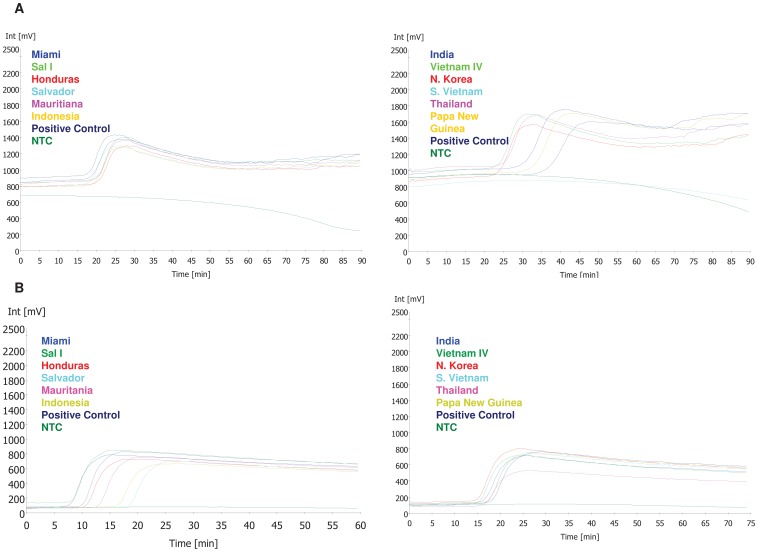
We found that HPLC purification of the primers provided better amplification plots with reduced background noise (data not shown). The time to amplification for all samples tested ranged from 18 to 49 minutes from the start of the reaction (Figure 1).
Figure 1. Amplification plots for P. vivax strains from different geographic locations using the r64 primer set with SYBR green (A) and SYTO-9 (B).
Amplification plots from P. vivax strains from Pakchong, Chesson and Panama are not shown. Amplification of the template DNA measured in millivolts (mV) on y-axis over time on x-axis. Any amplification observed after 60 minutes of test time was considered negative. A known P. vivax DNA sample was used as the positive control. NTC; No Template Control.
Specificity of P. vivax specific LAMP primers
We tested the novel primer sets on all known human malaria parasites (P. vivax, P. falciparum, P. malariae, P. ovale, and P. knowlesi). Only P. vivax was amplified (Table 2). In addition, no cross reactivity was observed with any of the tested simian malaria parasite species or with the malaria-negative human DNA control (Table 2).
Table 2. Specificity results of the r64 primer set when tested against human-infecting and simian-infecting malaria parasites.
| Human and simian malaria parasite species | RealAmp Result |
| P. falciparum | − |
| P. vivax | + |
| P. ovale | − |
| P. malariae | − |
| P. knowlesi | − |
| P. simiovale | − |
| P. inui | − |
| P. coatneyi | − |
| P. hylobati | − |
| P. fragile | − |
| P. gonderi | − |
| P. fieldi | − |
| P. cynomolgi | − |
| Malaria-negative human DNA | − |
All species were tested at least twice for reproducibility.
In order to validate the specificity of primer set r64, P. vivax strains from different geographic regions (Miami, El Salvador, Honduras, Mauritania, Indonesia, India, South Vietnam, North Korea, Thailand, New Guinea, Panama, Chesson, and Pakchong) were tested. We were able to amplify all of the different strains using our standard protocol (Figure 1 and data not shown).
Limits of detection, sensitivity and specificity of LAMP primers in clinical samples
Using serial dilutions of two P. vivax strains from Brazil and India, the limit of detection of r64 primer set was determined to be 125 parasites/µl and the limit of detection of previously published P. vivax LAMP primers (Han et al.) was determined to be between 250–500 parasites/µl. Clinical samples were used to determine primer sensitivity and specificity. The r64 primer set accurately detected 70/74 P. vivax-positive samples. No amplification was observed with 46 P. vivax-negative samples. Compared to a nested PCR reference, r64 sensitivity and specificity are 94.59% (95% CI: 87.48–98.26%) and 100% (95% CI: 90.40–100%) respectively (Table 3). This is a drastic improvement over the sensitivity of previously published P. vivax LAMP primers (36.11% (95% CI: 25.37–48.35%) with a specificity of 100% (95% CI: 90.40–100%)) in our hands.
Table 3. Sensitivity and specificity among clinical sample of r64 primer set and published P. vivax LAMP primers.
| Nested PCR (n) | Pvr64 | |
| Positive | Negative | |
| Positive (74) | 70 | 4 |
| Negative (46) | 0 | 46 |
| Sensitivity | 94.59% (95% CI: 87.48–98.26%) | |
| Specificity | 100% (95% CI: 90.40–100%) | |
Nested PCR using published 18S rRNA primers was used as the gold standard. All samples were tested at least twice for reproducibility.
SYBR Green versus SYTO-9
The specificity and sensitivity of the P. vivax r64 assay with SYTO-9 fluorescent dye was found to be the same as that of the assay with SYBR Green. In addition, the SYTO-9 amplification plots had lower background noise and in some cases, a better distinction from baseline fluorescence than the SYBR Green assays (Figure 1).
Discussion
Though the global distribution and disease burden of P. vivax is considerable, current molecular diagnostic methods that accurately detect P. vivax are limited. The RealAmp platform is a potential tool for using molecular diagnostic techniques in the field. The assay is simple, and provides rapid results through real-time data collection. This combination is ideal for developing countries that may not have the capacity and infrastructure to maintain PCR-based assays at every health care facility. The objective of this study was to design and test the utility of a novel set of LAMP P. vivax-specific primers using different target DNA sequences. The previously described genome sequence data-mining methods [20], adapted in this study, have proved to be useful in designing novel specific primers for P. vivax detection. Recent studies have developed new P. falciparum diagnostic LAMP primers based on mitochondrial DNA [26] and the apical membrane antigen-1 (AMA-1) gene sequence [27] was used to amplify P. knowlesi DNA. These new targets will help optimize the RealAmp assay for accurate detection of all malaria parasite species.
We have noted previously that Plasmodium subtelomeric regions harbor repetitive regions that are well suited to species-specific diagnostic targets [20], [28]. As the majority of Pvr64 copies are not assembled onto chromosomes, and the short assemblies containing these copies do not contain telltale signs of subtelomeric regions such as vir genes, we cannot be certain whether or not Pvr64 is a subtelomeric repeat. However, one chromosomal copy does occur ∼ 500 kb interior to the end of the assembly, arguably outside of the nebulously defined “subtelomeric” region. Though Pvr64 has some overlapping regions in other Plasmodium parasites such as P. falciparum and P. knowlesi, the primers developed in this study did not show any cross reactivity with other examined malaria parasites. While Pvr64 is similar to the 18S rRNA target in that it is relatively low-copy and not species-specific, it offers a superior, more sensitive RealAmp target than the 18S rRNA gene family, perhaps due to its comparatively high degree of conservation within the P. vivax genome.
The r64 primer set has shown reasonable sensitivity and specificity in detecting P. vivax among the clinical isolates used, accurately detecting 70 out of the 74 positive samples. It is possible that the four P. vivax samples that this primer set missed had low parasite density. Unfortunately, we were not able to determine this since we did not have any parasitemia data for the samples used in this study. Since P. vivax is genetically similar to several simian malaria parasites, we also tested the r64 primer set on eight simian parasites and observed no cross-reactivity. These primers were also able to amplify 13 P. vivax strains from different geographic locations. It is therefore likely that this assay will be able to detect P. vivax strains in malaria-endemic regions around the world.
Attempts to use published P. vivax LAMP primers by Han et al. in the RealAmp platform failed to yield consistent results, and the sensitivity was not as high as previously reported [15]. Possible explanations for this observation is the fact that different nested-PCR methods were used as the gold standard in calculating the sensitivity and specificity and that different detection methods were used in these studies. We also tested another published set of P. vivax LAMP primers by Tao et al [29]. However, these primers cross reacted with the other human-infecting Plasmodium species (P. falciparum, P. ovale and P. malariae), hence were not considered for further evaluation.
In some studies, SYBR Green was shown to have inhibitory effects on DNA amplification when used in high concentrations [22], [23]. We did not observe any inhibitory effect with SYBR Green in our assays. However, we obtained better amplification plots with SYTO-9 with lower background noise and in some cases, a better distinction from baseline fluorescence. This suggests that SYTO-9 is a better dye than SYBR Green for RealAmp assays and is possibly due to perfect spectral match with the detector. This is an important component of the assay as it will reduce misclassification of cases and non-cases by the users in the field.
One of the limitations of this primer set is its low sensitivity. Our data suggests that the limit of detection of the r64 primer set is around 125 parasites/µL when a quantified standard was used. This level of sensitivity might be sufficient for case management of malaria but not for the detection of sub-clinical malaria cases which is important during the pre-elimination phase of malaria. Efforts are underway to improve these limits of detection to enable detection of lower parasitemia. However, compared to nested PCR as a gold standard, this method performed reasonably well with approximately 95% sensitivity in detecting the clinical samples in a single assay.
As previously described, the RealAmp assay [19] has several advantages that make it a good choice for field-based molecular diagnosis of malaria. However, some hurdles need to be overcome for the RealAmp assay, and any molecular assay, to be truly field-usable. For example, a major caveat of molecular assays is the sample preparation phase; many assays require that the template DNA is purified and isolated. While the use of boiled whole blood was shown to give decent results with the RealAmp assay [19], additional studies are required to further simplify this step. In addition, the cold chain requirement for reagents used by many molecular assays needs to be addressed. Studies are underway to investigate if the RealAmp reagents can be combined into a “kit” which would further simplify the protocol, reduce the number of steps for the end user, and minimize chances of user-introduced errors in performing this assay. Lyophilized reagents that can be reconstituted at a later time would reduce preparation time and address the issue of reagent storage without a cold chain supply.
In conclusion, this study demonstrates that the P. vivax LAMP primers developed here have the ability to accurately detect P. vivax infections on the RealAmp platform. Furthermore, the previously described genome sequence data-mining methods have proved to be useful in designing primers not just for conventional PCR [20] but also for LAMP assays. This technique offers great potential in identifying novel primer sets for detection of other Plasmodium species.
Supporting Information
Comparison of the three sequences. The Pvr64 segment used to design the novel P. vivax primers was aligned with the P. knowlesi and P. falciparum sequences. The locations of the six LAMP primers are underlined (italicized sequences indicate location of the complementary sequences). Nucleotide differences found in the P. knowlesi (highlighted in green) and P. falciparum (highlighted in yellow) are shown.
(DOCX)
Acknowledgments
The authors would like to thank Jatan Patel and Zubin Mehta for their bioinformatics assistance in screening putative target sequences.
Funding Statement
This work was supported by a CDC-UGA seed grant (OPHR #8212) awarded to JCK and VU. This study was supported in part by resources and technical expertise from the University of Georgia Research Computing Center, a partnership between the Office of the Vice President for Research and the Office of the Chief Information Officer and CDC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2011) The top 10 causes of death.
- 2.WHO (2011) World Malaria Report.
- 3. Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, et al. (2009) Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis 9: 555–566. [DOI] [PubMed] [Google Scholar]
- 4. Galinski MR, Barnwell JW (2008) Plasmodium vivax: who cares? Malar J 7 Suppl 1S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, et al. (2007) Vivax malaria: neglected and not benign. Am J Trop Med Hyg 77: 79–87. [PMC free article] [PubMed] [Google Scholar]
- 6. Kochar DK, Saxena V, Singh N, Kochar SK, Kumar SV, et al. (2005) Plasmodium vivax malaria. Emerg Infect Dis 11: 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Genton B, D'Acremont V, Rare L, Baea K, Reeder JC, et al. (2008) Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med 5: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mayxay M, Pukrittayakamee S, Newton PN, White NJ (2004) Mixed-species malaria infections in humans. Trends Parasitol 20: 233–240. [DOI] [PubMed] [Google Scholar]
- 9.WHO (2010) Guidelines for the treatment of malaria.
- 10. Mens P, Spieker N, Omar S, Heijnen M, Schallig H, et al. (2007) Is molecular biology the best alternative for diagnosis of malaria to microscopy? A comparison between microscopy, antigen detection and molecular tests in rural Kenya and urban Tanzania. Trop Med Int Health 12: 238–244. [DOI] [PubMed] [Google Scholar]
- 11. Moody A (2002) Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, et al. (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16: 223–229. [DOI] [PubMed] [Google Scholar]
- 14. Chen JH, Lu F, Lim CS, Kim JY, Ahn HJ, et al. (2010) Detection of Plasmodium vivax infection in the Republic of Korea by loop-mediated isothermal amplification (LAMP). Acta Trop 113: 61–65. [DOI] [PubMed] [Google Scholar]
- 15. Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, et al. (2007) Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paris DH, Imwong M, Faiz AM, Hasan M, Yunus EB, et al. (2007) Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am J Trop Med Hyg 77: 972–976. [PubMed] [Google Scholar]
- 17. Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, et al. (2006) Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52: 303–306. [DOI] [PubMed] [Google Scholar]
- 18. Poschl B, Waneesorn J, Thekisoe O, Chutipongvivate S, Karanis P (2010) Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am J Trop Med Hyg 83: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, et al. (2010) Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One 5: e13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demas A, Oberstaller J, Debarry J, Lucchi NW, Srinivasamoorthy G, et al. (2011) Applied genomics: data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA. J Clin Microbiol 49: 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, et al. (1999) A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg 60: 687–692. [DOI] [PubMed] [Google Scholar]
- 22. Gudnason H, Dufva M, Bang DD, Wolff A (2007) Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res 35: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eischeid AC (2011) SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res Notes 4: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. (2008) Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, et al. (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res 37: D539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, et al. (2010) Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol 48: 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lau YL, Fong MY, Mahmud R, Chang PY, Palaeya V, et al. (2011) Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J 10: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lucchi NW, Poorak M, Oberstaller J, Debarry J, Srinivasamoorthy G, et al. (2012) A New Single-Step PCR Assay for the Detection of the Zoonotic Malaria Parasite Plasmodium knowlesi. PLoS One 7: e31848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tao ZY, Zhou HY, Xia H, Xu S, Zhu HW, et al. (2011) Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors 4: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the three sequences. The Pvr64 segment used to design the novel P. vivax primers was aligned with the P. knowlesi and P. falciparum sequences. The locations of the six LAMP primers are underlined (italicized sequences indicate location of the complementary sequences). Nucleotide differences found in the P. knowlesi (highlighted in green) and P. falciparum (highlighted in yellow) are shown.
(DOCX)