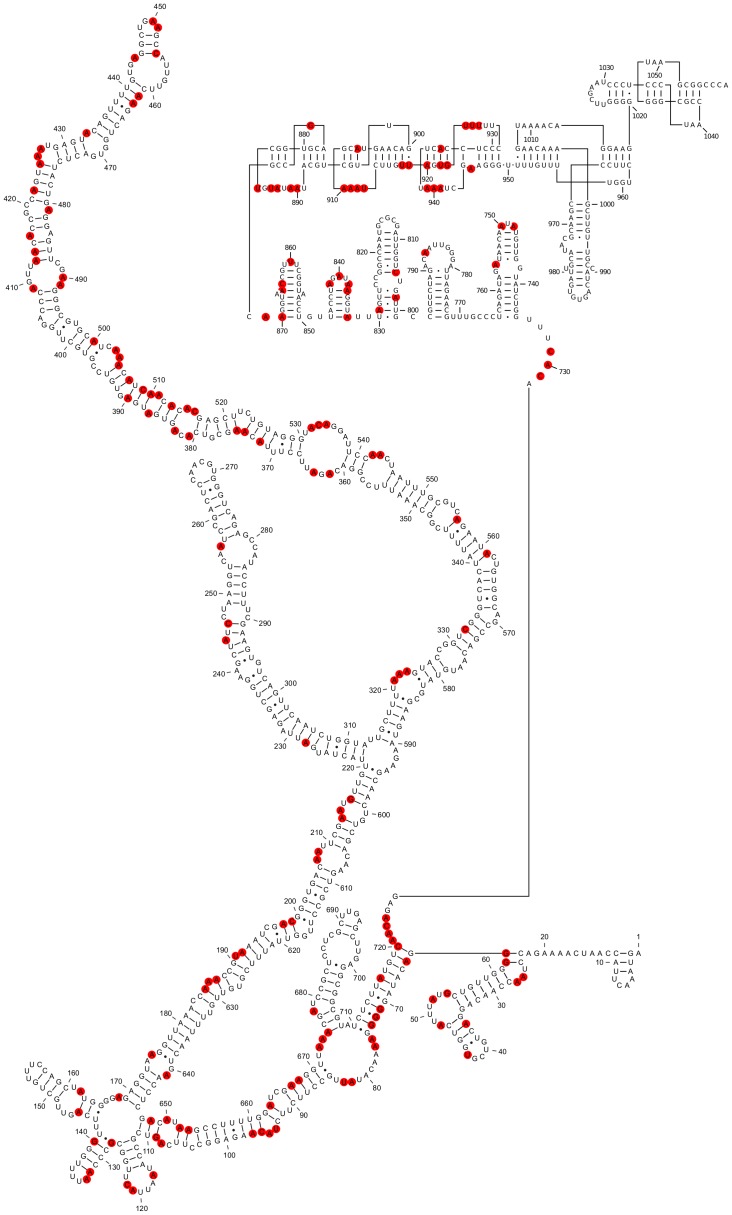
Figure 8. Mapping the chemical probing data from Schroeder et al. [7] onto the SHAPE-restrained secondary structure of in vitro transcribed STMV RNA.
Red circles indicate nucleotides modified by DMS, kethoxal, or CMCT. The data do not appear to clearly rule out the proposed secondary structure of residues 1–730. A substantial number of the modifications occur in predicted loops, bulges, and single-stranded regions (67 out of 119 hits). Many of the reactive base-paired nucleotides are in A-U or G-U base pairs immediately adjacent to a predicted bulge loop (e.g., 128, 185, 187, 192, 213, 413–414, 556, 561, 652–653, 663, 675), while others (382–390 and 503–515) are in a predicted stem that has two bulges and has no run of more than three consecutive base pairs, so it should be prone to fraying.