Figure 1.
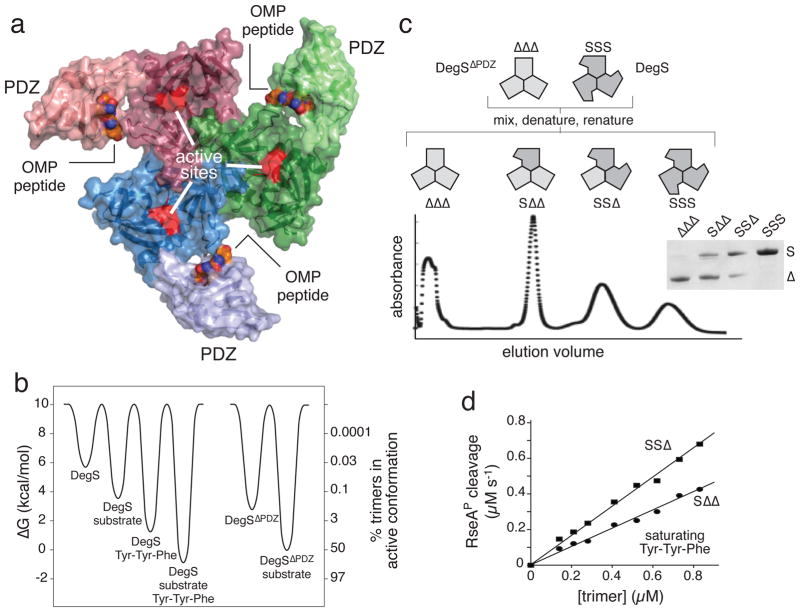
DegS and hybrid trimers containing mixtures of full length (S) and ΔPDZ (Δ) subunits. (a) Each subunit of the DegS trimer contains a trypsin-like protease domain (darker color; active site shown in red) and a peripheral PDZ domain (lighter color). Contacts between protease domains stabilize the trimer. OMP peptides (CPK representation) bind to the PDZ domains. The structure shown (3gcn)27 is the proteolytically active conformation. (b) Free-energy differences for DegS and DegSΔPDZ with and without saturating RseAP substrate and/or OMP peptide (Tyr-Tyr-Phe) were calculated (ΔG = − RT ln ([active]/[inactive]) from values taken from references 24 and 26. The right axis shows the percentage of active trimers for each molecular species. DegSΔPDZ has no binding site for OMP peptide. (c) The denaturation-renaturation protocol used to generate hybrid trimers is shown in the upper portion of the panel. The lower portion shows separation of hybrid trimers by ion-exchange chromatography. The inset shows SDS-PAGE of the four major peaks (see Supplementary Fig. 2 for complete gel). (d) The steady-state rate of RseAP (200 μM) cleavage by the SSΔ or SΔΔ variants showed a linear dependence (R2 > 0.98) on enzyme concentration. Reactions contained 150 μM Tyr-Tyr-Phe tripeptide.