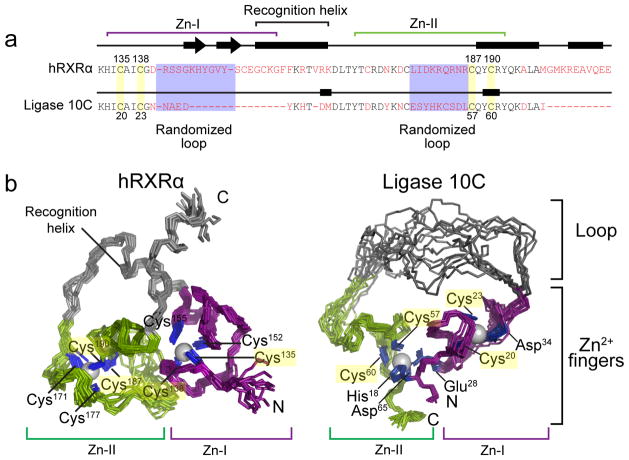
Figure 1. Changes in primary sequence and three-dimensional structure upon directed evolution of the hRXRα scaffold to the ligase enzyme 10C.
(a) Comparison of the primary sequences of hRXRα16 (residues 132–208) and the artificially evolved ligase (residues 17–68). The two zinc finger regions are highlighted with purple and green brackets. The red letters denote residues that are not conserved between the two sequences. (b) Three-dimensional structure of hRXRα16 and NMR ensemble of ligase 10C (for clarity, flexible termini are not shown). Although both proteins contain two zinc fingers, the overall structures are substantially different. Only two zinc-coordinating cysteines of each zinc finger in hRXRα are still coordinating zinc in ligase 10C (highlighted in yellow, see also Fig. 1a) while all other ligands differ in the two structures. In contrast to hRXRα zinc finger Zn-II in ligase 10C comprises residues of both N- and C-terminal sequence, imposing a cyclic structure to the enzyme. Residues involved in zinc coordination are labeled and shown in blue. Note that the new ligase lost both helical domains of the DNA binding domain (grey in left structure), replacing the recognition helix with a long unstructured loop (grey in right structure).