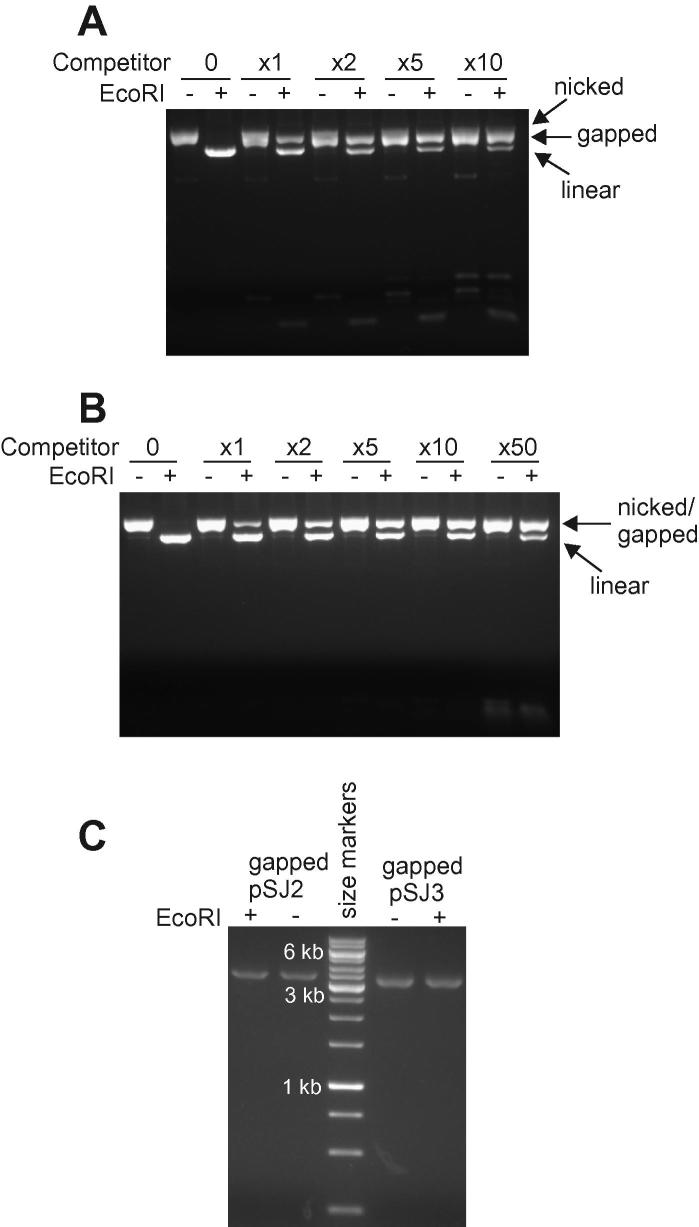
Fig.2.
Preparation of gapped pSJ2 and pSJ3. (A) Gel electrophoretic analysis of the generation of gapped pSJ2. The nicked form (previously prepared by reaction of pSJ2 with Nt.BbvCI) is converted to the desired gapped product by heat/cool cycles with a 288-base competitor oligodeoxynucleotide (excess used indicated above the gel lanes). The nicked starting material and the gapped product are poorly resolved. However, EcoRI converts the nicked plasmid (but not the gapped plasmid) to the well-separated linear form, enabling analysis of the progress of the gapping reactions. (B) Gel electrophoretic analysis of the preparation of gapped pSJ3. The nicked form (previously prepared by reaction of pSJ3 with Nt.Bpu10I) can be converted to the desired gapped product using heat/cool cycles with two 80-base competitors (excess used indicated above the gel lanes). The starting nicked plasmid and the gapped product are not resolved, and EcoRI digestion, which converts remaining nicked pSJ3 to the linear form but does not act on the gapped form, is required for analysis. (C) Gel electrophoresis of pSJ2 and pSJ3 following purification using BND–cellulose. Analyses were carried out with or without pretreatment of EcoRI to fully control for any contaminating nicked plasmid. The size marker is a GeneRuler 1-kb ladder (Fermentas), with the three intense bands being 1-, 3-, and 6-kb products, as indicated on the gel.