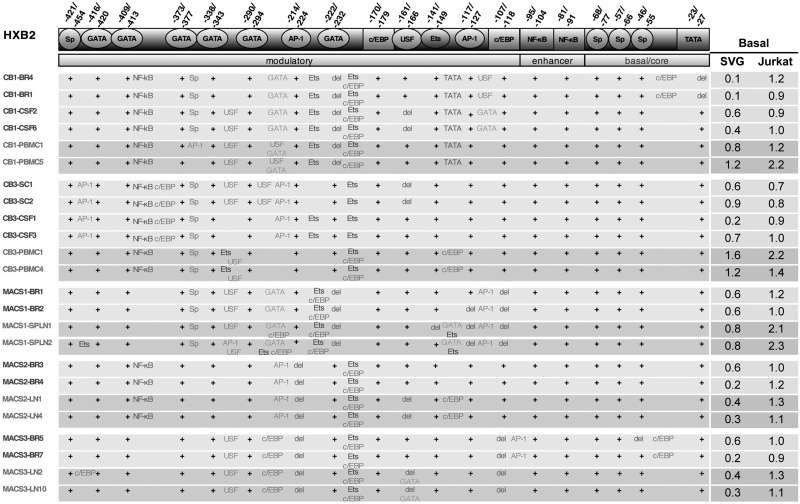
FIG. 2.
Transcription factor motif prediction of patient LTR sequences. LTR sequences were analyzed using “TF search” for the presence of a variety of transcription factor binding sites (TFBS) as compared to the HXB2 LTR. Sites with 75% homology to consensus or above are shown. A plus indicates that the respective site in HXB2 is retained in the patient LTR. Where deletion of a site occurs, this is represented by “del.” Text within the table represents additional TFBS. Clone names for CNS-derived LTRs are shown in black and non-CNS-derived LTRs are shown in gray. The corresponding relative basal transcriptional activities of each LTR in SVG and Jurkat cells are shown on the right. Basal values are shown relative to the wild-type NL4-3 LTR.