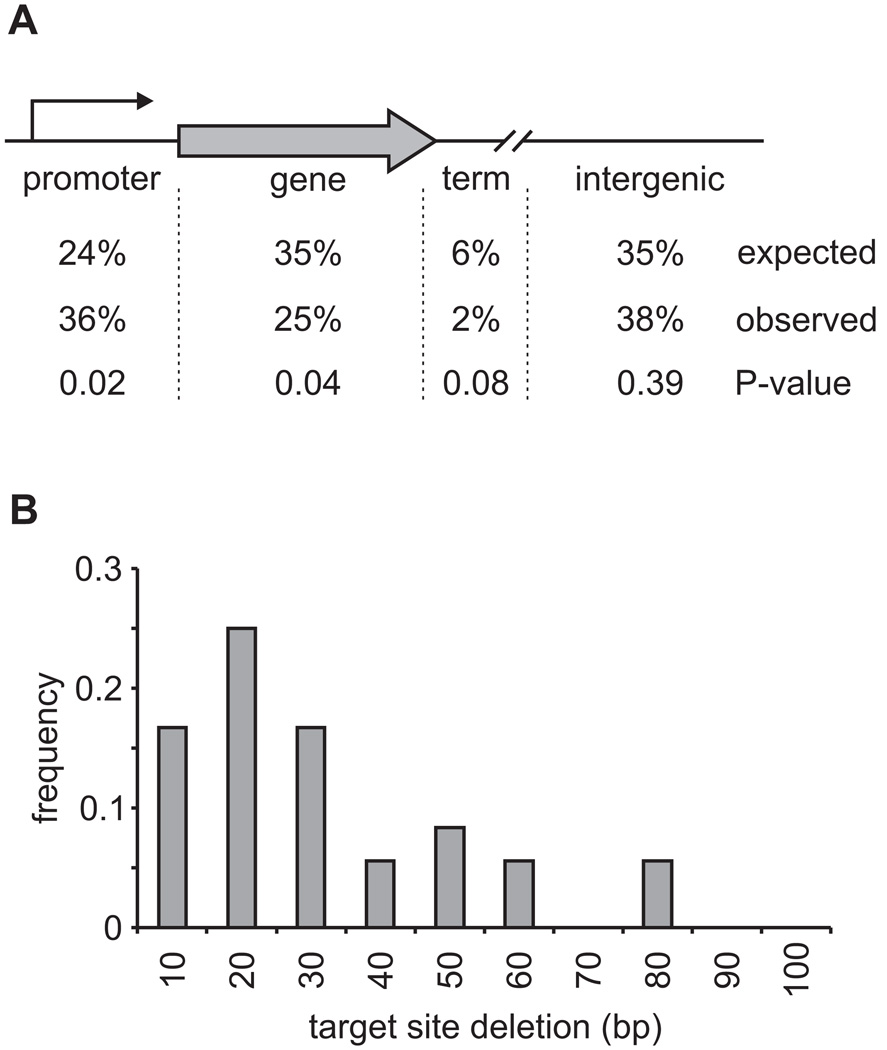
Figure 2. T-DNA integrations in Histoplasma are biased to promoter regions and are characterized by small genome deletions at the integration site.
(A) Distribution of T-DNA integration locations relative to genes in the Histoplasma genome. Values indicate the percentage of T-DNA integrations in promoters (within 800 bases upstream of the initiation codon), genes (exons + introns), terminator regions (up to 200 base pairs downstream of the termination codon), and intergenic locations. LB and RB T-DNA ends were considered individually (n=117 total) for the observed percentages. The expected proportion of the genome encoding gene sequences was derived from the base pairs encoding all predicted genes in the Histoplasma genome. Total promoter and terminator regions were estimated using the 11751 predicted genes. Remaining base pairs after calculation of promoters, genes, and terminators were considered intergenic. P-values were calculated using Chi-square analysis and differences were considered significant for P < 0.05. (B) Size distribution of deleted genome sequence associated with T-DNA integrations. Histogram shows the range and extent of genome deletions in base pairs (bp) for 86% of T-DNA integrations for which paired LB and RB-flanking sequence matched within the same contig (n=33). Five larger deletions of 292, 306, 386, 751, and 3381 base pairs were also observed (15% of the total).