Figure 2.
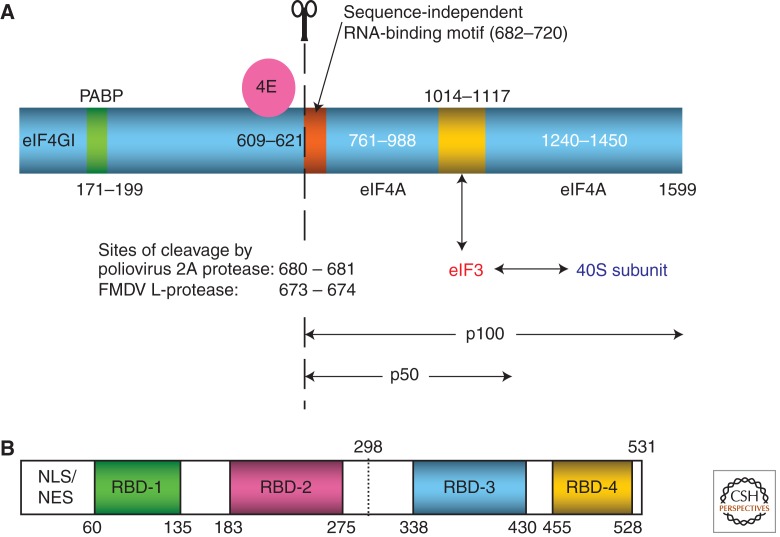
Domain structures of eIF4GI and polypyrimidine tract binding protein (PTB). (A) Domain structure of the longest isoform (1599 amino acids) of eIF4GI (blue) with associated eIF4E (magenta), showing the interaction sites of poly(A) binding protein (PABP) in green, and eIF3 (gold). The two sites of potential interaction with eIF4A are shown, although there is usually only a single bound eIF4A. The sites at which eIF4GI is cleaved by poliovirus 2A protease and FMDV L-protease are shown; the 2A cleavage site defines the amino termini of the carboxy-terminal two-thirds fragment (p100) of eIF4GI, and the central one-third domain (p50). The sequence-independent RNA-binding motif at the amino terminus of p100 (and p50) is highlighted in orange; this motif is necessary for scanning, but is not required for internal initiation on the EMCV IRES (Ali and Jackson 2001; Prévôt et al. 2003). (B) Domain structure of PTB-1. The amino-terminal ∼55 amino acid residues have nuclear import and export signals, but play no part in RNA-binding. The positions of the 4 RBDs (RNA-binding domains) are shown. RBDs-2 and -3 are longer than the other two RBDs because their RNA-binding surface has an additional β-strand. The linkers between the RBDs are flexible except for that between RBDs-3 and -4, which interact with each other in a back-to-back configuration, and act as a coordinated pair (Oberstrass et al. 2005). PTB-2 and PTB-4 differ from the canonical PTB-1 in having inserts (arising from alternative splicing) of 19 or 26 amino acids, respectively, at residue 298 of PTB-1.