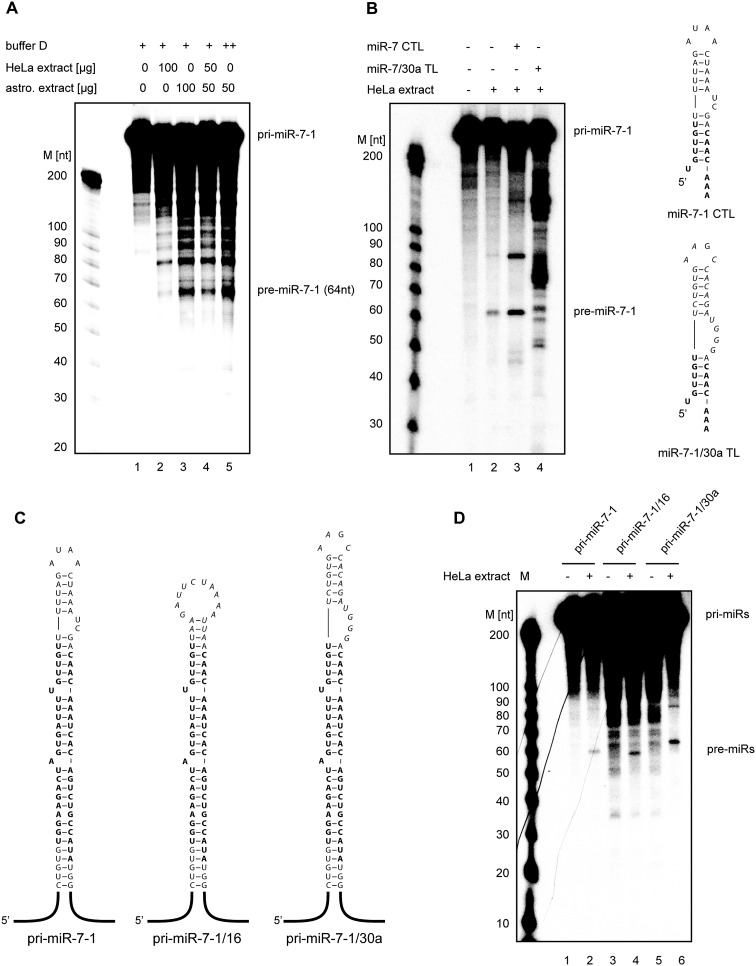
Figure 2.
Processing of pri-miR-7-1 is inhibited by nonneural CTL-binding factors. (A) In vitro processing of pri-miR-7-1 in HeLa and astrocytoma 1321N1 cell extracts. Radiolabeled primary miR-7-1 transcripts (50 × 103 cpm [counts per minute], ∼20 pmol, 665 nM) were incubated in the presence of 50% (w/v) total HeLa extract (lane 2), 50% (w/v) astrocytoma 1321N1 extract (lane 3), 25% (w/v) HeLa extract plus 25% (w/v) astrocytoma 1321N1 extract (lane 4), or 25% (w/v) astrocytoma 1321N1 extract (lane 5). Lane 1 shows the negative control with no extract added. Products were analyzed on an 8% polyacrylamide gel. (M) RNA size marker. (B) In vitro processing of pri-miR-7-1 is derepressed in the presence of unlabeled truncated wild-type pri-miR-7-1. pri-miR-7-1 was incubated with HeLa extracts (lane 2) with 1 nmol of miR-7-1 CTL (lane 3) or 1 nmol of pri-miR-7-1/30a CTL (lane 4). Lane 1 shows the negative control with no extract added. Analysis was performed as described in A. Schematic of the RNA secondary structure of the wild-type miR-7-1 CTL and mutant miR-7-1/30a TL is shown on the right. (C) Predicted secondary structures of wild-type pri-miR-7-1 and CTL mutants. The wild-type terminal loop sequence was replaced with the pri-miR-16 or pri-miR-30a TLs, resulting in pri-miR-7-1/16 and pri-miR-7-1/30a hybrids, respectively. (D) The processing of pri-miR-7-1 is inhibited by sequences present in its terminal loop. Radiolabeled pri-miR-7-1 (lane 2), pri-miR-7-1/16 (lane 4), and pri-miR-7-1/30a (lane 6) transcripts (50 × 103 cpm, 20 pmol, 665 nM) were incubated in the presence of 50% (w/v) total HeLa extract. Lanes 1, 3, and 5 show negative controls with no extract added. Products were analyzed on an 8% polyacrylamide gel. (M) RNA size marker.