Abstract
Purpose of review
Molecular genetics is revolutionizing the diagnosis and treatment of inherited eye diseases. The National Eye Institute of the National Institutes of Health (NIH), in an effort to facilitate future basic and clinical research in inherited eye disease, created The National Ophthalmic Disease Genotyping and Phenotyping Network (eyeGENE). This review describes the process and utility of the eyeGENE program as it relates to ophthalmic clinical practice.
Recent findings
Over the last few years, genetic testing of specific genes associated with inherited eye conditions is becoming the standard practice. Vision research and human clinical trials relying on molecular genetic testing of individuals with inherited eye conditions are becoming more common. Eye healthcare professionals must consider the options to assist patients in obtaining genetic testing results and locating trials or studies that may have benefit.
Summary
eyeGENE is a DNA repository and patient registry for inherited eye diseases coupled to phenotypic descriptors and molecular genetic information. Through eyeGENE, healthcare professionals throughout the United States and Canada can obtain Clinical Laboratory Improvement Amendments-certified clinical molecular genetic results on their patients. Researchers may request access to a de-identified database of phenotype and genotype information about eyeGENE participants and DNA aliquots for their research studies. eyeGENE also offers participants the option of being included in a patient registry, whereby they may be re-contacted if an approved clinical study for which they might qualify is offered.
Keywords: biorepository, clinical trials, eyeGENE, genetic testing, patient registry
INTRODUCTION
The last decade has been exciting for those interested in vision research, with significant advances in the development of potential treatments for gene-based causes of vision loss. A search of www.clinicialtrials.gov reveals the trials actively recruiting patients for the treatment of Leber congenital amaurosis (LCA), choroideremia, Stargardt disease, and age-related macular degeneration, all gene-associated eye conditions. In 2008, three independent groups published the promising results of gene replacement therapy for LCA caused by mutations in the RPE65 gene [1–3]. Animal studies of gene replacement therapy have also shown promise in stabilizing or slowing photoreceptor loss and improving visual function in disorders due to mutations in other genes such as BBS4 [4▪▪], GUCY2D [5], and CNGB3 [6▪▪]. Additionally, recent advances in stem cell therapy for retinal diseases have shown promise in animal models. As discussed in detail in a recent Current Opinion in Ophthalmology review [7], retinal pigmented epithelium and photoreceptors have been successfully derived from human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). Transplantation into rodent models has been shown to restore some visual function [8–11]. Additional strategies, including the use of small molecules [12▪▪], small interfering RNAs [13], and retinal prostheses [14], are also being tested. Although these strategies are in various stages of development, individuals with inherited eye conditions and the vision research community are encouraged by the potential treatment implications of these strategies for many previously untreatable conditions.
These successes are accompanied by significant scientific and practical hurdles. For example, many forms of inherited eye disease are uncommon. So, identifying patients for study and treatment trials can be difficult. Many clinical trials testing gene-based strategies may require a patient to know the specific genetic cause of their condition. Genetic testing can be expensive and not fully covered by health insurance. In addition, although new information is continually being discovered on genetic eye diseases, there is still much that is unknown. In order to overcome these hurdles, the National Eye Institute (NEI) created eyeGENE as a resource to support research into the genetic causes of inherited eye disease and to aid clinicians and researchers across the USA and Canada in their care of and research on patients with these diseases, while simultaneously helping patients obtain Clinical Laboratory Improvement Amendments (CLIA)-certified molecular genetic testing through their clinician.
eyeGENE
In 2003, the NEI convened a series of meetings with ophthalmic geneticists, genetic counselors, regulatory specialists, and members of the vision community from around the world. Participants advocated for the establishment of a community resource to bring together individual genetic testing laboratories for the common goals of patient care, education, and research. Recommendations were also made for a resource to be made available for vision community researchers to obtain DNA specimens that were reliably collected and stored from individuals with well characterized, inherited eye conditions. At that time, clinical testing facilities were difficult to locate and only a few were available. Members of the ophthalmology community expressed a need for a centralized resource to obtain genetic testing for their patients with inherited eye conditions. The NEI sought to leverage a collaborative framework that would allow for clinical information to be linked to genetic results, DNA samples, and a patient registry.
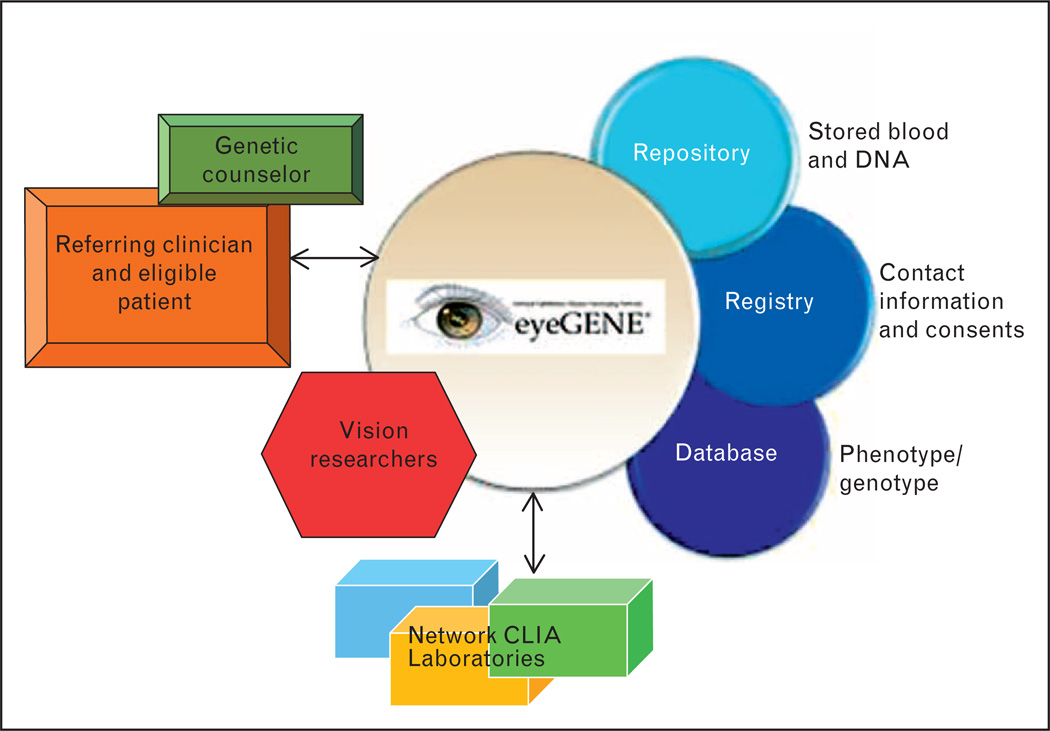
The following years realized the development of The National Ophthalmic Genotyping and Phenotyping Network (ClinicalTrials.gov Identifier NCT00378742, NIH protocol #06-EI-0236), also known as eyeGENE. A model partnership, eyeGENE is a collaboration between the federal government, eye healthcare providers, CLIA approved molecular diagnostic laboratories, private industry, and the vision research community. The eyeGENE network consists of CLIA-certified clinical molecular diagnostic testing facilities, a central Coordinating Center, a biorepository of DNA samples, a research database, referring clinical organizations, and a patient registry (Fig. 1). Patients enrolled in eyeGENE agree to allow the NEI to store their DNA and distribute de-identified aliquots to vision researchers. Patients also have the option to be included in the eyeGENE Registry.
FIGURE 1.
Organization of the eyeGENE Network. Source: eyeGENE Working Group.
The eyeGENE network responds to the needs of three broad groups: patients, clinicians, and researchers. In brief, eye healthcare providers submit patient blood samples for molecular DNA diagnoses, along with phenotypic information. Patients receive genetic testing results and can choose to contribute their information to a confidential, de-identified registry from which they could be selected for future clinical trials. Researchers may apply for access to de-identified patient samples and the de-identified database for curated genotype and phenotype information and patient samples. Once their application for access is approved, researchers can access the secure de-identified database at no charge. Depending on the study proposed by the researcher, approval from their Institutional Review Board (IRB) may be required. As the approved research study is completed, data is returned to the eyeGENE Coordinating Center and entered back into the eyeGENE database to ensure a more robust community resource. Approved research studies may result in opportunities for collaboration between the researcher and the referring eye healthcare provider. The network framework (described in more detail in the following paragraphs) can be viewed as a genetic collaboration in which different components serve to expand research while providing a more complete diagnostic report for clinicians and patients.
The referring clinician identifies the eligible participant and, after obtaining the consent of the participant, submits a blood specimen, along with signed consent forms (see below), family history, and eye exam information for the patient. Once the eyeGENE Coordinating Center reviews the information and forms for accuracy and completeness, the DNA is extracted from the blood specimen. A coded portion of the sample is sent to a network CLIA laboratory. Upon completion of testing, results are de-coded by the Coordinating Center and returned to the referring clinician. If results are negative, that is, no disease-causing mutation is found, the samples will be re-tested at a later time as new technology and genetic information is available to make re-testing a sample a reasonable option. In this situation, if a disease-causing mutation is found through subsequent testing, a new report will be sent to the referring clinician with this information. Also, given today’s ever changing genetic landscape, if the interpretation of a mutation is changed in the scientific literature, for example, a nondeleterious mutation is now thought to be pathogenic or vice versa, an amended report will be sent to the referring clinician. Vision researchers may access eyeGENE to obtain de-identified phenotype and genotype data, DNA samples, or request that eyeGENE staff contact patients to inform them of possible recruitment for clinical trials. When a patient is contacted, the referring clinician is also informed. Researchers request access through a formal process, and the research proposal is reviewed by the eyeGENE Resource Access Subcommittee (a sub-committee of the eyeGENE Steering Committee, which oversees the eyeGENE program).
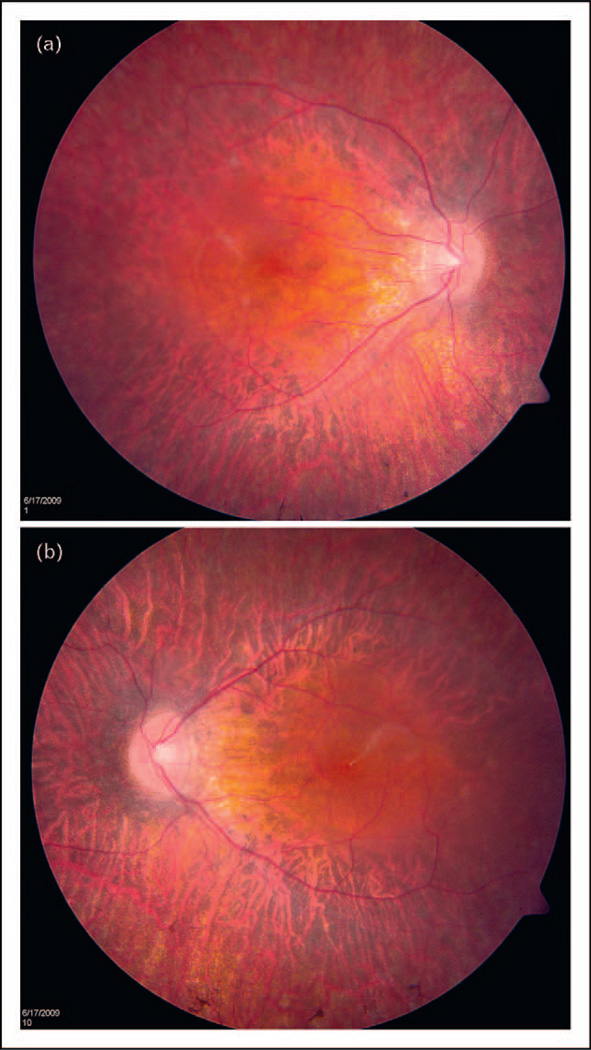
The eyeGENE study was piloted in the fall of 2006 when the first patient sample was received. The program has now surpassed 3700 enrollments. Not only has the eyeGENE program been a success based upon participation statistics, but also there are numerous cases in which the program has resulted in meaningful results for patients. In one case (Fig. 2), a 13-year-old boy was enrolled in the program with a clinical diagnosis of retinitis pigmentosa. There was no known family history of the disease. His visual acuity was 20/40 in each eye, and the dark-adapted electroretinogram (ERG) was extinguished, while the light-adapted/flicker ERG was greatly reduced. The patient showed typical intraretinal pigment spicules, but vascular attenuation was not present. He did not suffer from hearing loss. Because of data presented in literature, estimating that approximately 30% of sporadic retinitis pigmentosa is caused by mutations in the RPGR gene [15], and accompanying clinical data, this patient was screened for mutations in RPGR and found to have a nucleotide duplication, causing a 5-amino acid frameshift in the protein (Exon 3, c.246dupA, p.Ala83fs*). Because this individual presented as a sporadic case, where the inheritance pattern could be due to autosomal recessive alleles, an X-linked condition, or a new dominant mutation, testing established a mode of inheritance, prompted examination of the patient’s mother, and modified genetic counseling.
FIGURE 2.
Color fundus photographs of 13-year-old boy with retinitis pigmentosa. (a) Right eye. (b) Left eye. Source: Dr Elias Traboulsi and the Cole Eye Institute.
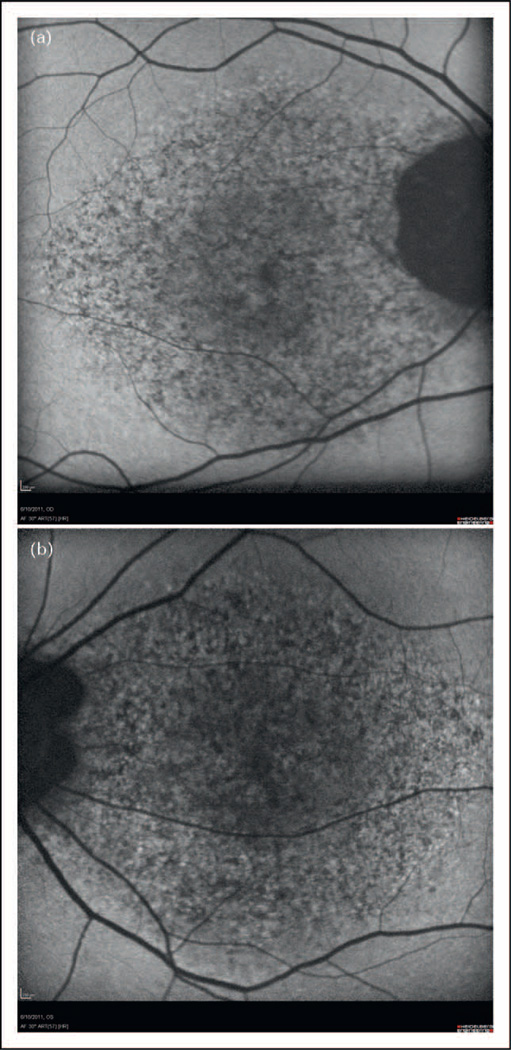
In a second case (Fig. 3), a 49-year-old man was enrolled in eyeGENE, having been diagnosed with Stargardt disease at age 32. The inheritance pattern was thought to be autosomal dominant because of an extensive family history reported as ‘macular dystrophy’. The clinician recorded the patient’s best corrected visual acuity as 20/100 right eye and 20/25 left eye, with a central scotoma and foveal atrophy bilaterally. A ‘questionable’ dark choroid by fluorescein angiogram was noted and the full-field ERG was within normal limits. This patient was initially tested through eyeGENE for mutations in the ELOVL4 gene, but had negative results. The patient was not tested for mutations in ABCA4, as dominant inheritance was indicated. Nearly 3 years later, a secondary diagnosis of pattern dystrophy was added for this patient. Upon a second evaluation at 52 years old, the visual acuity was measured at 20/80 right eye and 20/30 left eye. This patient was now noted to have a depigmented lesion in the fovea, central hypofluorescence surrounded by hyperfluorescence by fundus autofluorescence bilaterally. The full-field ERG was still within normal limits. The patient was tested for mutations in the PRPH2 (RDS) gene and found to have a point mutation in PRPH2 (RDS), (Exon 1, c.514C>T, p.Arg172Trp), a gene associated with pattern dystrophy. The confirmation of the diagnosis helped the referring physician make a firm gene-based diagnosis and provide counseling in a case in which the presentation of the macular dystrophy was somewhat unclear.
FIGURE 3.
Fundus autofluorescence of 52-year-old man (a) Right eye. (b) Left eye. Source: Dr Jacque Duncan and the University of California, San Francisco.
ENROLLING PATIENTS IN eyeGENE
Any board-certified healthcare provider within the USA and Canada may enrol patients in eyeGENE through registration of their clinic in a secure online database. The clinic may enrol up to 10 patients per calendar year under the purview of the NIH/NEI IRB; however, enrolment of more than 10 patients requires individual or commercial IRB approval. Once registered, the clinic must enter the participant’s clinical information into the secure database and submit their blood sample to the eyeGENE Coordinating Center for extraction and genetic testing. Results of the genetic testing are reported directly back to the referring healthcare provider to be shared and explained to the participant.
Registering for use and institutional review board approval
The eyeGENE program was designed to allow eye healthcare providers throughout the USA and Canada, whether or not they are affiliated with larger public institutes or universities, to enroll their patients. Because of the rarity of several conditions included in eyeGENE (Table 1), it is not likely that most clinicians will enroll more than a few patients in their entire career. Patients referred by registered providers enrolling less than 10 patients per year may be enrolled as offsite participants of the NIH using the NIH/NEI-approved consent forms and are not required to have their own IRB approval. However, a referring eye healthcare provider should always check with their local IRB, if available, for their local requirements and regulations. The NEI forms are available for download from the eyeGENE database. In these cases, a member of the NEI eyeGENE Coordinating Center staff will call the patient to verify that the consents have been reviewed with them. For sites enrolling over 10 patients per calendar year, eyeGENE requires local or commercial IRB approval. At these sites, the Principal Investigator is responsible for administering patient consents using the locally approved consent forms provided by the IRB. Additionally, all referring providers agree to assist the patient in obtaining both pregenetic and postgenetic testing counseling regardless of whether or not an organization seeks independent IRB approval. If the provider does not have access to a genetic counselor, they may refer to the National Society of Genetic Counselors (www.nsgc.org) to find resources in their area.
Table 1.
Genes and diseases currently being tested by the eyeGENE network
| Diagnoses eligible for inclusion | Genes that may be tested |
|---|---|
| Albinism | Recessive TYR, OCA2, TYRP1, SLC45A1 X-linked GPR143 (OA1) |
| Aniridia and other developmental eye anomalies | PAX6, WT1, DCDC1, ELP4 |
| Axenfeld–Rieger syndrome | PITX2, FOXC1 |
| Best disease | BEST1 |
| Bietti’s crystalline corneoretinal dystrophy | CYP4V2 |
| Choroideremia | CHM |
| Chronic progressive external ophthalmoplegia/Kearns–Sayre syndrome, mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes, myoclonic epilepsy associated with ragged red fibers, neuropathy, ataxia, and retinitis pigmentosa |
Mitochondrial gene panel |
| Cone rod dystrophy | ABCA4, RPGR, CRX, GUCY2D (codon R838) |
| Congenital cranial dysinnervation diseases | KIF21A, CHN1, SALL4, TUBB3 |
| Congenital stationary night blindness | RHO, NYX |
| Corneal dystrophy | TGFBI, KRT3, KRT12 |
| Doyne honeycomb dystrophy | EFEMP1 |
| Familial exudative vitreoretinopathy | FZD4, LRP5, NDP, TSPAN12 |
| Glaucoma | CYP1B1, OPTN, MYOC |
| Hermansky–Pudlak syndrome | HPS1 and HPS3a |
| Infantile neuroaxonal dystrophy | PLA2G6 |
| Juvenile X-linked retinoschisis | RS1 |
| Leber hereditary optic neuropathy (LHON) | LHON panel (MT-ND4, MT-ND1, MT-ND6/ mutations 11778G>A, 3460G>A, 14484T>C, and 14459G>A) |
| Lowe syndrome | OCRL |
| Microphthalmia and anophthalmia | SIX6, SOX2, OTX2, CHX10 |
| Optic atrophy type 1 | OPA1 |
| Pantothenate kinase-associated neuropathy | PANK2 |
| Pattern dystrophy | RDS |
| Retinitis pigmentosa and retinal degenerations | Dominant (panelb including RHO, PRPH2, RP1, IMPDH1, PRPF8, NR2E3, PRPF3, TOPORS, PRPF31, RP1, KLHL7), CA4, CRB1, CTRP5 X-linked RPGR, RP2 |
| Retinoblastoma | RB1 |
| Sorsby fundus dystrophy | TIMP3 |
| Stargardt disease | ABCA4, ELOVL4, RDS |
Updated 15 September 2011.
In individuals of Puerto Rican decent, test screens for a 16-bp duplication in HPS1 and a 3.9 kb deletion in HPS3. Ashkenazi Jewish individuals will be tested for IVS5 splice site mutations in HPS3 only. HPS samples from individuals outside of these categories will not be tested at this time.
Not all genes sequenced in full and not all are available outside of panel.
Any board certified or credentialed eye healthcare provider (e.g. ophthalmologist, optometrist, neurologist, geneticist and genetic counselor) practicing within the USA and its protectorates or Canada, who is caring for a patient with a candidate genetic eye condition, may register as long as she or he can address the phenotypic and clinical criteria questions related to the patient’s presentation. A registered clinical organization can be a sole practitioner in a private practice or a medium to large practice or university with multiple practitioners. There are detailed online instructions at http://www.nei.nih.gov/resources/eyegene/professionals.asp or the referring clinician may opt to speak with Coordinating Center staff on the phone. To register, a user must fill in the required information on the secure database site https://nationaleyegene.nei.nih.gov. The eyeGENE Coordinating Center, located on the NIH main campus in Bethesda, Maryland, will review the information provided and approve the organization’s use of the program. After registering, the referring clinician may designate other staff (e.g. administrators, nurses, etc.) as ‘users’ who are authorized to input clinical criteria and other patient data when samples are submitted. Each enrolled patient must read and sign the research consent form and a DNA diagnostic consent form. A list of the responsibilities of referring clinicians is shown in the list below. Responsibilities of referring clinician are as follows:
register as an eyeGENE user;
identify eligible patients;
discuss eyeGENE Project with prospective participants;
administer consents/assent;
arrange for genetic counseling;
enter family history and eye exam information along with photos and electrophysiology to the eyeGENE database;
assist patient in arranging for a blood draw to be shipped to the eyeGENE Coordinating Center;
share any genetic testing results with the patient and genetic counselor.
There are currently over 30 conditions listed as eligible for enrollment (Table 1). The list includes more common diseases like retinitis pigmentosa, Stargardt disease, and albinism along with rare diagnoses like pantothenate kinase-associated neuropathy (PKAN) and congenital cranial dysinnervation disorders. It is important to note that there are some conditions (Table 1), for which eyeGENE allows patients to be enrolled, but does not currently offer testing. For example, eyeGENE enrols patients with retinitis pigmentosa and cone rod dystrophy with unknown or recessive inheritance, but does not offer clinical testing at this time. Separate research efforts are ongoing to develop multigene tests for these conditions, and these samples may be tested on a research basis. If a positive result, that is, a disease-causing mutation, is found through research efforts, eyeGENE will confirm the research result in a CLIA-certified diagnostic laboratory so that the patient and the referring eye healthcare provider can receive the results. Once a test is available for a diagnosis pending in the repository, samples with that diagnosis will automatically be tested.
Enrolling a patient
Once a clinical organization’s registration is approved, the first step to enrol a patient in eyeGENE is for the clinical organization to enter the patient’s data into the secure online database. This is true whether the practitioner enrolls less than 10 cases per year or 100. The user must enter information regarding a patient’s family history and submit answers to a series of questions related to the suspected condition. This data, along with the family history, is critical to the research efforts of the network, and, subsequently, is reviewed and curated by a team of eyeGENE ophthalmologists to ensure accuracy and value for research. At any step in the process, the referring clinician can ask to speak with an NEI ophthalmologist who is reviewing the patient file. The entered patient data is also used by the Coordinating Center to determine which genetic test(s) to perform and in what order. The referring clinician is encouraged to provide additional evidence of the diagnosis when enrolling the patient that may lead to specific genes being tested. eyeGENE may request additional patient data if it is not initially provided. Common requests are for photography, ERG data, and visual fields. The eyeGENE database also allows the referring clinician and their approved users to upload these documents directly to the database. Additionally, the referring clinician is able to select, on a patient-by-patient basis, an option to be contacted for possible scientific collaborations if a vision researcher would like additional information for a more in-depth study. Researchers utilizing eyeGENE data and materials are required to adhere to NIH authorship polices. This allows the referring eye healthcare provider not only an opportunity for scientific collaboration, but also the ability to actively participate and contribute to patient research studies if she or he desires.
After entering the information in the database, the referring provider will assist the patient in contributing a blood sample to the eyeGENE Coordinating Center. One of the major goals for eyeGENE is to maintain a biorepository of DNA samples from patients with inherited eye conditions. Therefore, the draw volume is more than traditional fee-for-service genetic testing facilities. Patients of age 16 and older are required to provide 24–30 ml of whole blood, while children of age 8–15 should provide 10–15 ml and younger children should give 7–10 ml. In some cases, multiple blood draws may be necessary. eyeGENE does not charge the referring clinician or the patient for DNA storage or genetic testing of the sample regardless of the number of genes tested, but the patients or their referring providers are responsible for the costs of phlebotomy, eye exams and any testing or photography performed, blood shipment, and fees associated with genetic counseling.
Once the clinical information is reviewed and found to substantiate the diagnosis and is sufficient for research, the eyeGENE laboratory will ship a portion of the stored DNA sample to a network CLIA-certified laboratory. The sample is coded with a unique identifier and once the test results are available, the Coordinating Center will link that code back to the patient. The CLIA laboratory does not have access to the patient’s name. The report will be reviewed by the eyeGENE Coordinating Center and the results sent back to the referring clinician. Results are also available for access by the referring clinician on the eyeGENE database with their user login. Users only have access to patients referred from their own organization or practice. Turnaround time for the receipt of molecular diagnostic results varies and often depends on the gene(s) tested and availability of a test. The average turnaround time is between 4 months and 1 year. The remaining DNA extracted from the blood samples and de-identified data for patients enrolled in eyeGENE is available to vision researchers who have received the approval for a research study through the eyeGENE Resource Access Subcommittee.
Through eyeGENE, patients are provided an avenue to aid in research related to their eye condition and may choose to be contacted for recruitment to clinical studies. Both the clinical providers and the patients are integral in providing an immensely valuable resource of linked phenotype–genotype data; a pool of highly characterized, recruitment eligible research subjects; and de-identified DNA samples to the research community.
Although eyeGENE offers benefits to many groups, there can also be associated risks for patients. It is important to remember that genetic testing may provide information about how an eye condition is passed on within a family and this can be a source of stress for some families. Also, even though a federal antidiscrimination law (Genetic Information Nondiscrimination Act of 2008; GINA; P.L. 110–233, 122 Stat. 881) is in place, workplace and insurance discrimination may occur if genetic information is disclosed. These risks are included in the eyeGENE consent forms and should be discussed with each patient upon consideration for enrollment.
NEW DEVELOPMENTS
eyeGENE began piloting the process for vision research access to the de-identified data, samples, and recruitment from the patient registry in August 2010 (NEI Protocol #10-EI-N164). Several studies are now underway and additional proposals are pending. The scope of studies is broad, showcasing the usefulness of the eyeGENE Network as a resource to the vision community. For example, one study is reviewing Stargardt clinical features and how they relate to the mutation information available for certain regions of the ATP-binding cassette, subfamily A, member 4 (ABCA4) gene. This study hopes to correlate genetic information of patients with Stargardt disease with the severity of phenotype. A second study is using DNA samples to determine if a gene of interest has implications in aniridia. Two other studies have found eyeGENE useful in recruitment to their independent vision-related protocols. These studies have expedited the ability to find patients not only with specific inherited conditions, but also with specific genetic profiles to recruit a specialized patient population conducive to generating induced pluripotent stem cells for Best disease.
Once research studies are completed, summary information will be posted for the general public.
eyeGENE plans to continue to expand both its recruitment efforts for specific inherited eye conditions as well as the number of approved research projects using the Network. eyeGENE plans to launch a new look to the eyeGENE website (http://nei.nih.gov/eyeGENE) and will include information tabs for patients, healthcare providers, and researchers. Summary data, and eventually results for approved studies, will be publicly available through this site. eyeGENE is also actively evaluating new technology for more complex genetic testing both within the NEI and in other laboratories and uses recent research data to test small samples of patients using very stringent criteria to test genetic associations with disease. To date, over 3000 gene tests have been performed through eyeGENE and nearly 2000 results have been returned to referring clinicians. It is easy to imagine that the role and value of eyeGENE will continue to grow as advances in targeted gene therapy develop. eyeGENE has become a significant resource to the vision community and, with support, will continue to advance the field well into the next decade.
CONCLUSION
The National Eye Institute’s eyeGENE program is a collaborative network that provides a unique opportunity for eye healthcare providers to assist patients with inherited eye conditions in participating in a research study facilitating a pooled resource for research interests. The framework of the program is instrumental in providing data and materials for research with little recruitment effort. Providers and patients may receive an added benefit of obtaining CLIA level genetic testing results.
KEY POINTS.
eyeGENE (National Institutes of Health/National Eye Institute) serves as a resource to clinicians, patients with inherited eye conditions, and the vision research community.
The eyeGENE Network includes a database of phenotypes linked to genotypes for patients with inherited eye conditions, a biorepository of corresponding DNA samples, and a de-identified patient registry that is accessible to the vision research community.
Eye care professionals may enroll eligible patients to eyeGENE to facilitate research in inherited eye conditions, become involved in potential collaborations, and aid in patient clinical care by obtaining genetic test results.
Individuals with inherited eye disease have the opportunity to receive molecular diagnostic testing and participate in vision research.
Acknowledgements
The authors would like to thank Dr Elias Traboulsi, Cole Eye Institute and Dr Jacque Duncan, University of California, San Francisco for providing the photos presented in the case studies as well as the eyeGENE participants for their valuable contributions to this research.
Funding: This study was supported by the Department of Health and Human Services/National Institutes of Health/National Eye Institute intramural program under eyeGENE – Protocol 06-EI-0236 which has been funded in part under Contract No. HHS-N-260-2007-00001-C.
Footnotes
Conflicts of interest
All authors work for the National Eye Institute at the National Institutes of Health and are involved in operations of eyeGENE.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (p. 451).
- 1.Maguire A, Simonelli F, Pierce E, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainbridge J, Ali R. Success in sight: the eyes have it! Ocular gene therapy trials for LCA look promising. Gene Therapy. 2008;15:1191–1192. doi: 10.1038/gt.2008.117. [DOI] [PubMed] [Google Scholar]
- 3.Hauswirth W, Aleman T, Kaushal S, et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simons D, Boye S, Hauswirth W, et al. Gene therapy prevents photoreceptor death and preserves retinal function in a Bardet–Biedl syndrome mouse model. Proc Natl Acad Sci USA. 2011;108:6276–6281. doi: 10.1073/pnas.1019222108. This study demonstrates successful gene therapy in Bbs4 knockout mice through a subretinal injection of the wild-type gene complement incorporated into a viral vector which subsequently restores localization of rhodopsin and visual function.
- 5.Boye S, Boye S, Pang J, et al. Functional and behavioral restoration of vision by gene therapy in the guanylate cyclase-1 (GC1) knockout mouse. PLoS One. 2010;5:e11306. doi: 10.1371/journal.pone.0011306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carvalho L, Xu J, Pearson R, et al. Long-term and age-dependent restoration of visual function in a mouse model of CNGB3-associated achromatopsia following Gene Therapy. Hum Mol Genet. 2011;20:3161–3175. doi: 10.1093/hmg/ddr218. This article shows that a treatment window exists in regards to age at gene supplementation treatment for the nonprogressive condition, achromotopsia caused by Cngb3, in mice with younger mice experiencing more effective response to treatment.
- 7.Tibbetts M, Samuel M, Chang T, et al. Stem cell therapy for retinal disease. Curr Opin Ophthalmol. 2012;23:226–234. doi: 10.1097/ICU.0b013e328352407d. [DOI] [PubMed] [Google Scholar]
- 8.Lu B, Malcuit C, Wang S, et al. Long-term safety and function of RPE from human embryonic stem cells in preclinical models of macular degeneration. Stem Cells. 2009;27:2126–2135. doi: 10.1002/stem.149. [DOI] [PubMed] [Google Scholar]
- 9.Lamba D, Gust J, Reh T. Transplantation of human embryonic stem cell derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell. 2009;4:73–79. doi: 10.1016/j.stem.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr A, Vugler A, Hikita S, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4:e8152. doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang K, Hopkins J, Heier J, et al. Ciliary neurotrophic factor delivered by encapsulated cell intraocular implants for treatment of geographic atrophy in age-related macular degeneration. Proc Natl Acad Sci USA. 2011;108:6241–6245. doi: 10.1073/pnas.1018987108. This clinical trial suggests that treatment through a retinal implant, which allows for the slow release of ciliary neurotrophic factor, in patients diagnosed with geographic atrophy, increases retinal thickness leading to a decrease in the rate of visual acuity loss compared with a sham treatment.
- 13.Hernan I, Gamundi M, Planas E, et al. Cellular expression and siRNA-mediated interference of rhodopsin cis-acting splicing mutants associated with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2011;52:3723–3729. doi: 10.1167/iovs.10-6933. [DOI] [PubMed] [Google Scholar]
- 14.Weiland J, Cho A, Humayun M. Retinal prostheses: current clinical results and future needs. Ophthalmology. 2011;118:2227–2237. doi: 10.1016/j.ophtha.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier V, Jambou M, Delphin N, et al. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype–phenotype correlations and impact on genetic counseling. Human Mutat. 2007;28:81–91. doi: 10.1002/humu.20417. [DOI] [PubMed] [Google Scholar]