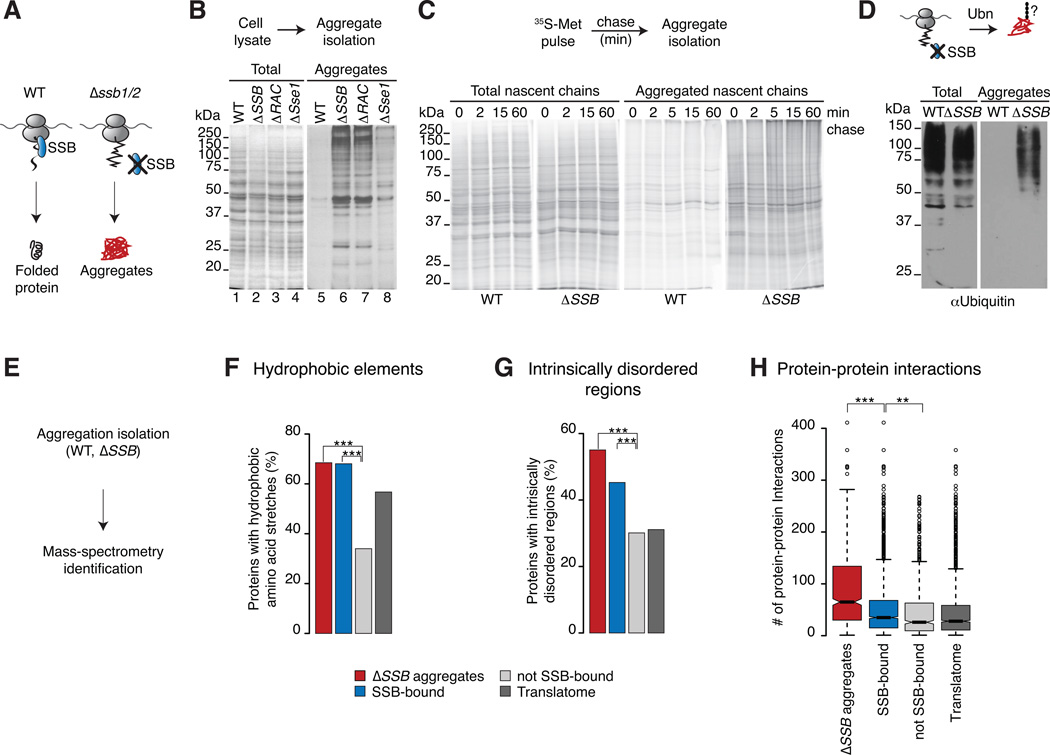
Figure 6. SSB maintains solubility of aggregation-prone nascent polypeptides.
(A) We hypothesize that SSB prevents aggregation of newly synthesized proteins. (B) Loss of SSB or RAC leads to widespread and loss of Sse1 leads to partial aggregation. The presence of insoluble proteins in WT, mutant cells was examined by SDS-PAGE and silver-staining (left panel: Totals; right panel: aggregates). (C) Loss of SSB leads to rapid aggregation of newly synthesized proteins. Newly made proteins of WT and ΔSSB cells were 35S-Met pulse labeled followed by a chase with cold methionine. Aggregates were isolated and analyzed by SDS-PAGE and autoradiography. (D) Proteins aggregated in ΔSSB cells are ubiquitylated as shown by SDS-PAGE and immunoblotting for Ub. (E) Global identification of aggregates in WT and ΔSSB cells and mass spectrometry. (F–G) Comparison of intrinsic properties between aggregates in ΔSSB cells, SSB substrates and non-SSB-bound proteins. The Translatome serves as reference. Only protein properties of cytosolic and nuclear localized proteins are shown. Proteins that aggregate in ΔSSB cells have similar intrinsic properties as cotranslational substrates of SSB. (H) Distinct enrichment of protein-protein interactions in protein aggregates of ΔSSB cells. *p ≤ 0.01; **p≤ 10−4; ***p ≤10−10. See also Figure S6.