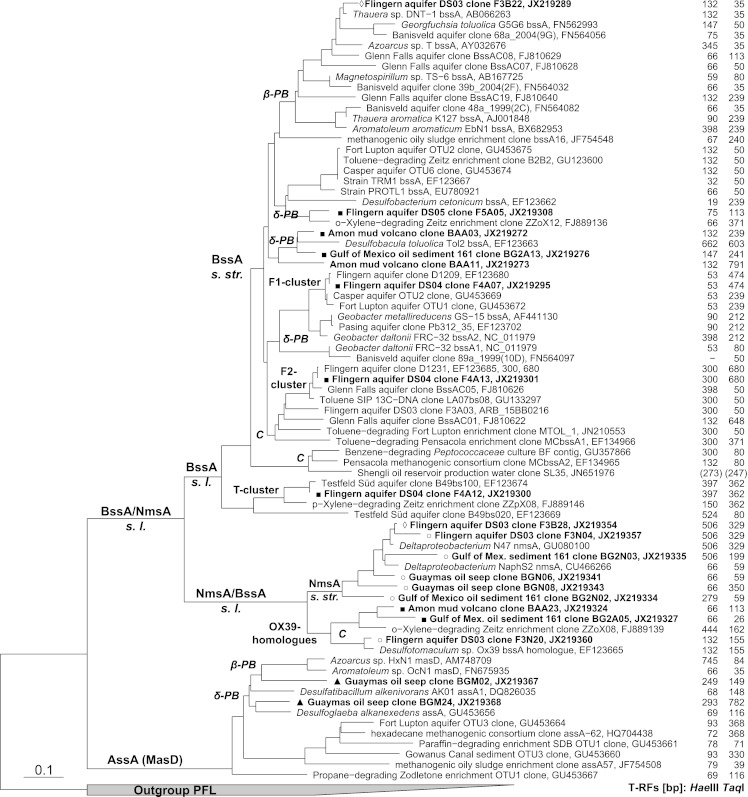
Fig 1.
Current overview of FAE gene phylogeny and affiliation of clones detected in this study. Sequences retrieved in this study are in bold and marked with different symbols according to the primer sets used: ◊, set FAE-B; ■, bssA at 52°C; ○, set FAE-N; ▲, set FAE-KM. Abbreviations of FAE lineages (as identified via enrichments or pure cultures): C, Clostridia; β-P, Betaproteobacteria; δ-P Deltaproteobacteria. The numbers to the right give the T-RFs predicted in silico for amplicons generated with the 8543r primer and a digest with HaeIII or TaqI. The T-RF sizes in parentheses are tentatively extrapolated, as some sequence entries end before the primer site. s. str. sensu stricto; s. l., sensu lato; PFL pyruvate formate lyase.