Fig 1.
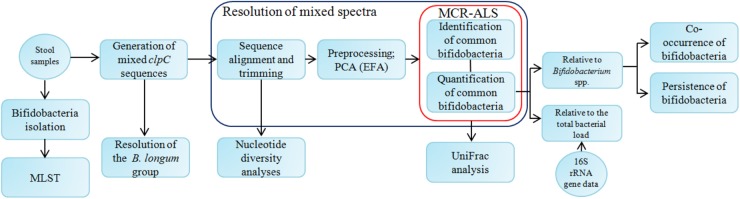
Flow diagram of the study. Boxes and circles represent processes and the data or materials obtained from them, respectively. At first, generated mixed spectra were trimmed to ensure that each spectrum brings an equal amount of information into the system. Then these spectra were analyzed using principal component analysis (PCA), evolving factor analysis (EFA), and multivariate curve resolution analysis with alternating least squares (MCR-ALS) to identify and quantify common bifidobacterial species in the data set. Quantification was performed relative to both the Bifidobacterium group and the total bacterial load. The B. longum group was resolved separately. Diversity was assessed using both taxon-based (UniFrac analysis based on MCR-ALS predictions) and taxon-independent (based on mixed spectra) techniques. The co-occurrence of various bifidobacteria, as well as their persistence over time, was also evaluated. Also, bifidobacteria were isolated from stool samples, and MLST analysis of several isolates was performed.
