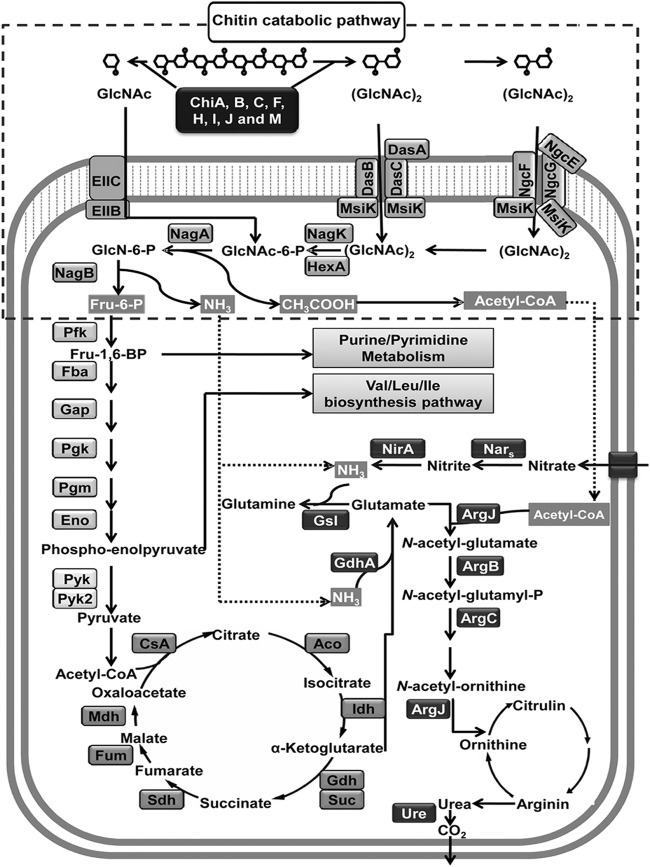
Fig 1.
Representation of the S. coelicolor A3(2) chitin metabolism, which was significantly influenced at the transcriptional level during growth in soil. Gene products for which expression was upregulated are shown in the metabolic pathways. Acetyl-CoA, acetyl coenzyme A; EIIC, enzyme IIC; EIIB, enzyme IIB; Pfk, 6-phosphofructokinase; Fba, fructose-bisphosphate aldolase; Gap, glyceraldehyde-3-phosphate dehydrogenase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, phosphopyruvate hydratase; Pyk, pyruvate kinase; CsA, citrate synthase A; Aco, aconitate hydratase; Idh, isocitrate dehydrogenase; Gdh, α-ketoglutarate dehydrogenase; Suc, succinyl-CoA synthetase; Sdh, succinate dehydrogenase; Fum, fumarate hydratase; Mdh, malate dehydrogenase; GsI, glutamin synthase I; ArgJ, N-acetylglutamate synthase J; ArgB, N-acetylglutamate kinase, ArgC, argininosuccinate synthase; Nar, nitrate reductase; Nir, nitrite reductase; Ure, urease.