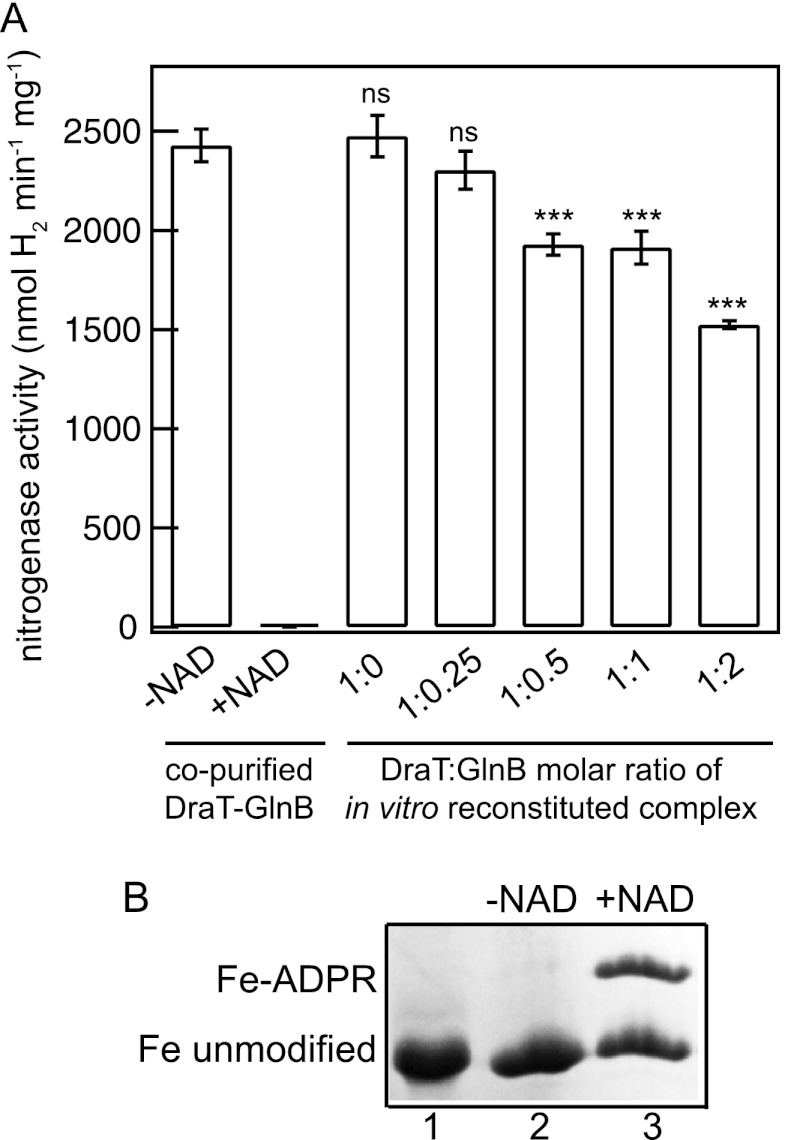
Fig 1.
Fe protein activity and modification after DraT-GlnB complex activity assay. (A) Fe protein activity was determined after incubation at different DraT/GlnB (monomer/trimer) ratios or in the presence of the DraT-GlnB complex obtained by copurification. The reaction mixtures for the DraT assay contained 2 mM NAD+; 1 mM MgCl2; 1 mM ADP; 100 μg of A. vinelandii Fe protein; and DraT, DraT-GlnB complex formed in vitro, or copurified complex and were incubated for 5 min at 130 rpm at 30°C. The reactions were performed using an Fe protein/DraT (dimer/monomer) molar ratio of 42:1. As a control, the reaction was performed in the presence of either copurified complex in the absence of NAD. The DraT-GlnB assay was stopped by reducing NAD+ using dithionite solution, the Fe protein activity was determined by adding 500 μg of the MoFe protein, and the hydrogen evolution was measured. ns, nonsignificant; ***, P < 0.001. The error bars represent the calculated standard deviations. (B) NAD+-dependent modification of A. vinelandii Fe protein catalyzed by the A. brasilense DraT-GlnB complex. Lane 1, control containing purified, unmodified A. vinelandii Fe protein. Lanes 2 and 3, purified A. vinelandii Fe protein was incubated with the A. brasilense copurified DraT-GlnB complex in the presence (lane 3) or absence (lane 2) of NAD+. The unmodified and ADP-ribosylated subunits of the Fe protein were separated using low-cross-link SDS-PAGE, and the gel was stained with Coomassie blue. The percentages of Fe-ADPR and unmodified Fe protein were quantified using Photoshop CS4, resulting in 42 and 58%, respectively.