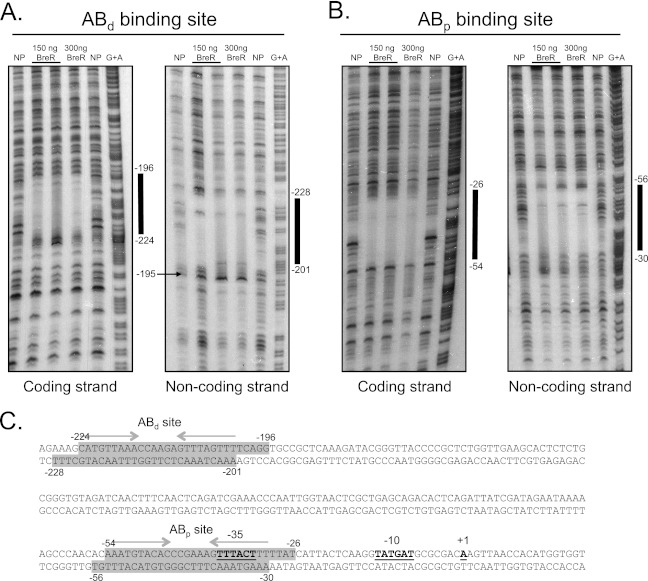
Fig 4.
DNase I footprinting analyses for BreR at the breAB promoter. The coding and noncoding strands of two radioactively 33P-end-labeled fragments of 230 bp (−327 to −97) (A) and 246 bp (−167 to +78) (B) were incubated with a 1:200 dilution of DNase I and no protein (NP; lanes 1 and 5), ∼150 ng BreR and a 1:200 DNase I dilution (lane 2) or a 1:400 DNase I dilution (lane 3), ∼300 ng BreR and a 1:200 dilution of DNase I (lane 4), and a G+A sequencing reaction (lane 6). The region of protection by BreR and a region of hypersensitivity induced by BreR are indicated by thick black lines and a black arrow, respectively. (C) Double-stranded DNA sequence of the breAB promoter. BreR-protected regions are shaded in gray for both strands. Regions of partial dyad symmetry, involved in DNA binding, are shown for the ABd and ABp binding sites with gray arrows. The transcriptional start site (+1) (17), ATG start codon, and putative −35 and −10 regions are highlighted in boldface and are underlined.