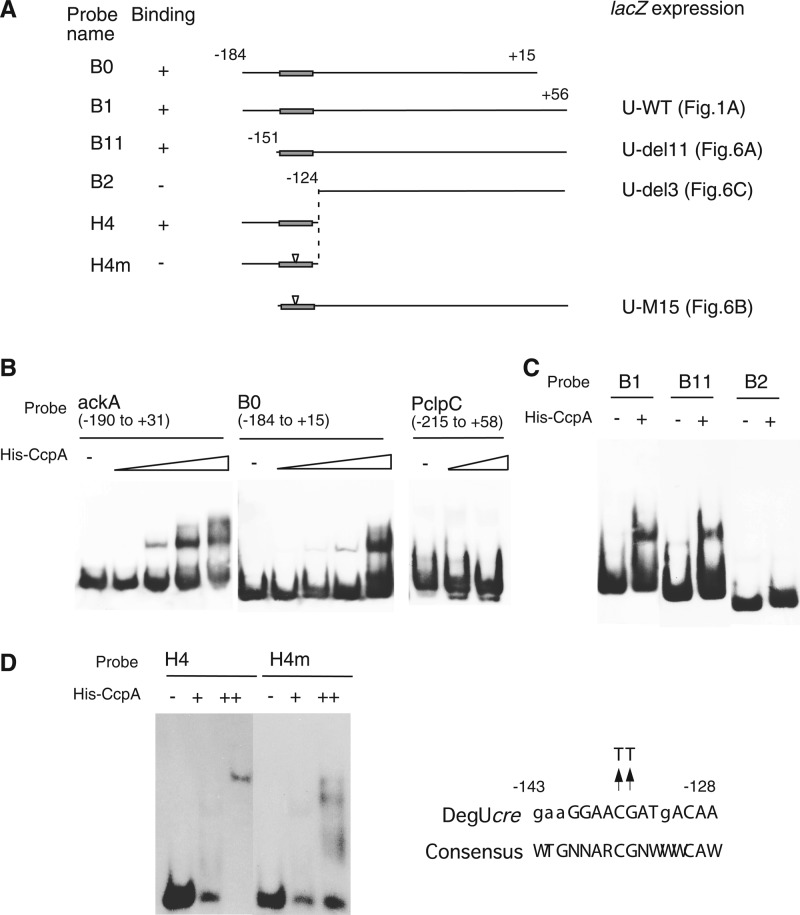
Fig 4.
Binding of His-CcpA to degU. Various probes were prepared by PCR amplification using the primers shown in Table S1 in the supplemental material. (A) Structures of probes. Solid rectangles on lines show cre sites. Numbers indicate nucleotide positions relative to the transcription start site. Relationships between probes and fusions with corresponding regions fused to lacZ are indicated. (B to D) Each reaction mixture contained poly(dI-dC) (1 μg [B], 0.7 μg [C], or 0 μg [D]) as a nonspecific competitor and probe. Samples were analyzed by nondenaturing polyacrylamide gel electrophoresis on 4.5% gels (B and C) or 11% gels (D). In each panel, all lanes shared the same gel electrophoresis conditions. For panel B, probes were incubated with increasing amounts of His-tagged CcpA (0, 12.5, 25, 50, and 100 nM for ackA and B0 and 0, 50, and 100 nM for PclpC). For panels C and D, probes were incubated with His-tagged CcpA (+, 75 nM; ++, 150 nM).