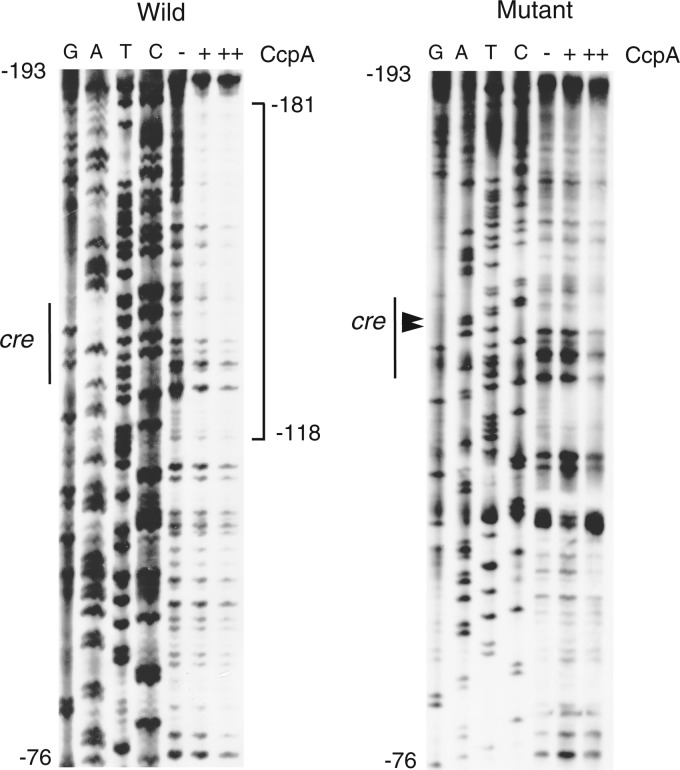
Fig 5.
Footprint analysis of His-CcpA binding to degU. To analyze the wild-type template strand, oligonucleotide pairs DegU-Pro-Biotin3/DegU-pIS-B1 and DegU-pIS-H3/DegU-pIS-B1 were used for PCR amplification to generate the probe and sequencing ladder template, respectively. To analyze the mutant template strand, the oligonucleotide pair DegU-Pro-Biotin3/DegU-pIS-B1 in addition to mCre-degU-F/mCre-degU-R was used to create a mutant probe for PCR-mediated mutagenesis (35). DegU-pIS-H3 and DegU-pIS-B1 were used for PCR amplification of the resultant mutant probe to generate the sequencing ladder template. The probe spanning positions −184 to +15 relative to the transcription start site, which carries the additional nine bases derived from the PCR primer, was incubated with His-CcpA (+, 75 nM; ++, 150 nM) in buffer containing poly(dI-dC) (0.5 μg) as a nonspecific competitor (the buffer had the same composition as the buffer used for EMSA) and subjected to DNase I cleavage. A sequencing ladder is shown, with G, A, T, and C lanes. The square bracket shows the protected region. The two mutated nucleotides are indicated by arrowheads.