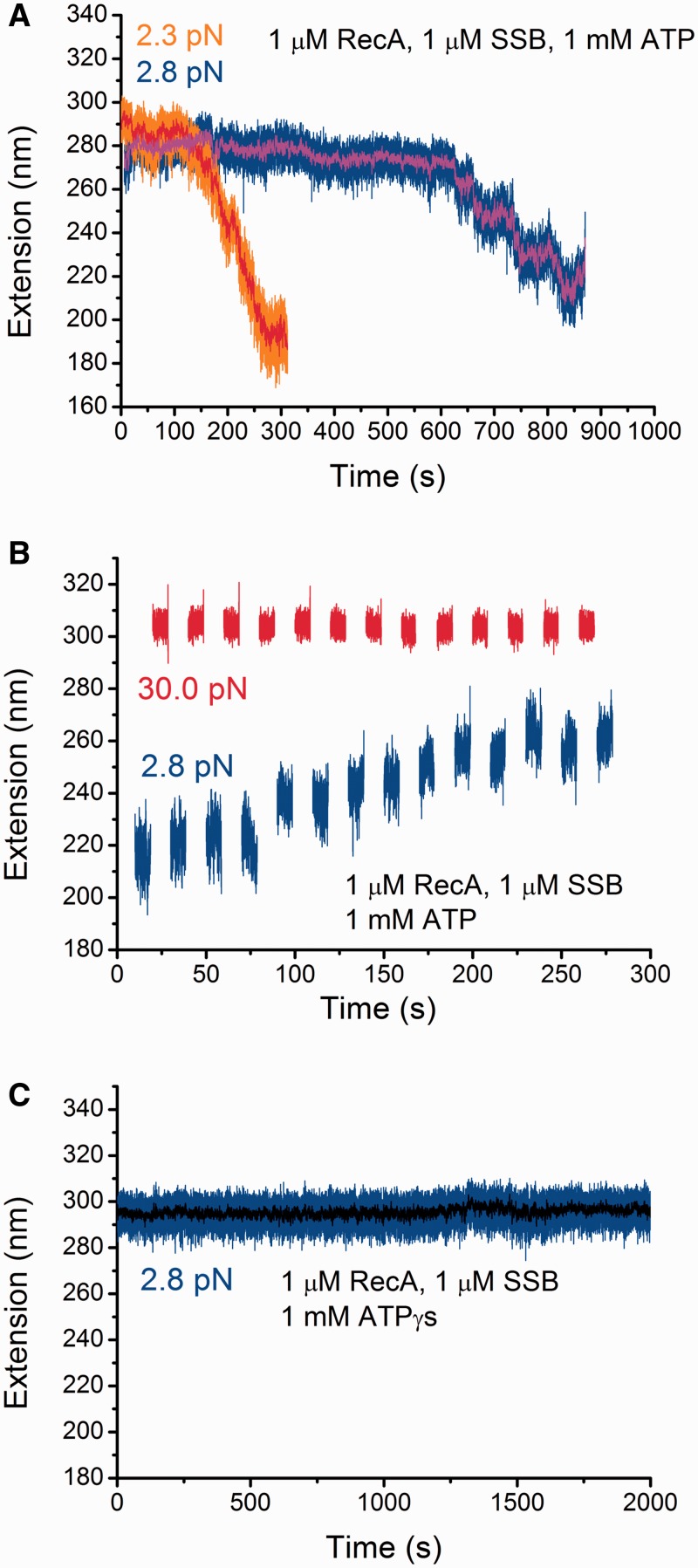
Figure 3.
Effects of SSB, ATP hydrolysis and force on the stability of pre-formed RecA nucleoprotein filaments. (A) Extension change of two different pre-formed RecA nucleoprotein filaments after introduction of protein mixture containing 1 µM RecA, 1 µM SSB, and 1 mM ATP, 1 × ATP regeneration system. Both exhibit a lag phase followed by a quick extension decreasing phase at low forces <3 pN. Raw data are in orange and blue, while the smoothed data by 40-points fast Fourier transformation are in red and magenta, respectively. (B) Extension change of a partially de-polymerized RecA nucleoprotein filament (the same ssDNA as indicated by blue in Figure 3A) during multiple cycles of jumping between a high force (∼30 pN) and a low force (∼2.8 pN). In general, these force-jump cycles lead to elongation of the ssDNA. (C) On the same ssDNA indicated by blue in Figure 3A, repeating the experiments described in Figure 3A but with a replacement of ATP with ATPγS. The result shows that the presence of SSB does not cause de-polymerization of the pre-formed RecA nucleoprotein filament with ATPγS.