Figure 2.
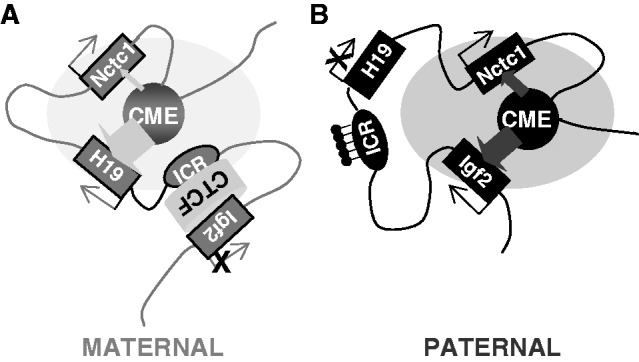
Parent-of-origin specific structures mediate gene expression at the Igf2/H19/Nctc1 imprinted locus in muscle cells. (A) On the maternal chromosome, a CTCF-dependent insulator organizes the DNA loops between distal cis-regulatory elements to favor H19 expression and to prevent interactions between the Igf2 promoters and the shared downstream CME. Recent work from Nativio et al. (17) demonstrates a critical importance for cohesin in establishing these maternal-specific chromosomal structures. Also, see Zhang et al. (25) for detailed mechanisms describing maternal ICR–CTCF–Igf2 interactions. (B) Paternal-specific methylation of CpGs within the ICR prevents CTCF binding, resulting in DNA loop structures that favor Igf2 promoter interactions with the shared enhancer. The loss of CTCF binding also results in a spread of DNA methylation and heterochromatin from the ICR into the adjacent H19 promoter region to H19 transcription. Here, we propose that Nctc1 levels are regulated by competition with H19 and Igf2 promoters for activation by the transcriptional complexes assembling at the shared enhancer. For simplicity, this model does not describe Igf2 differentially methylated regions (DMRs) located near the Igf2 promoters. DMR1 is an muscle-specific repressor of Igf2 expression that is required for complete full postnatal repression of both maternal and paternal chromosomes (62). DMR2 is a tissue non-specific positive regulatory element (63). In muscle, deletion of DMR2 results in modest decreases in Igf2. The participation of these two elements in DNA looping structures has been extensively analyzed (12,13,64) but to date, only in endodermal cells.
