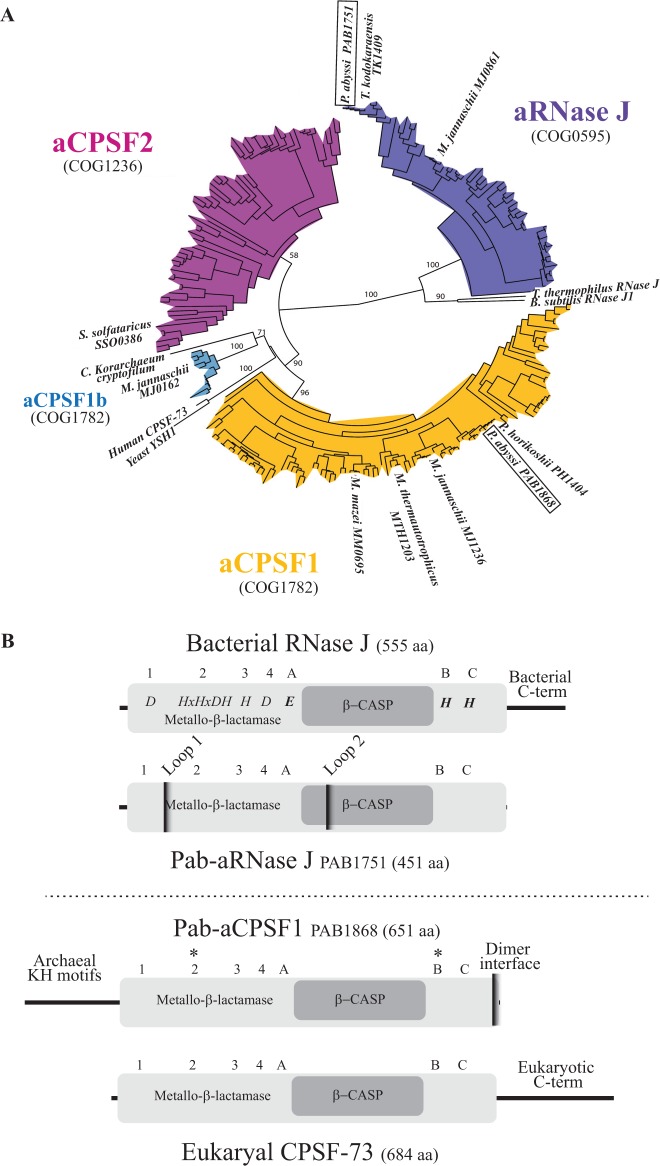
Figure 1.
The archaeal metallo-β-lactamase β-CASP protein family. (A) Phylogenetic tree of the four β-CASP OGs. The β-CASP proteins with reported structure or/and activity were added as landmarks: yeast and human CPSF 73 kD (10), M. thermautotrophicus MTH1203 (14), P. horikoshii PH1404 (16), M. mazei MM0695 (26), B. subtilis RNase J1 (48), Thermus thermophilus RNase J (21,49), P. abyssi PAB1751 (24), M. jannaschii MJ0861, MJ0162 and MJ1236 (15), S. solfataricus SSO0386 (25). The P. abyssi β-CASP members are framed. It should be mentioned that the landmarked Candidatus Korarchaeum cryptofilum β-CASP member is not considered as a CPSF1b fellow in view of its sequence. (B) Schematic representation of Pab-aRNAse J (PAB1751) and Pab-aCPSF1 (PAB1868), homologs of bacterial RNase J (Top) and eukaryal CPSF-73 (Bottom), respectively. The β-CASP and metallo-β-lactamase domains are highlighted in dark and light grey, respectively. The three β-CASP motifs (A-C), four metallo-β-lactamase motifs (1–4) and N- and C-terminus are indicated with respective features. Pab-aRNase J loop 1 and 2 insertions are indicated (24). The asterisks (*) indicate the position of conserved amino acids that have been mutated in this study, H261A and H594A.