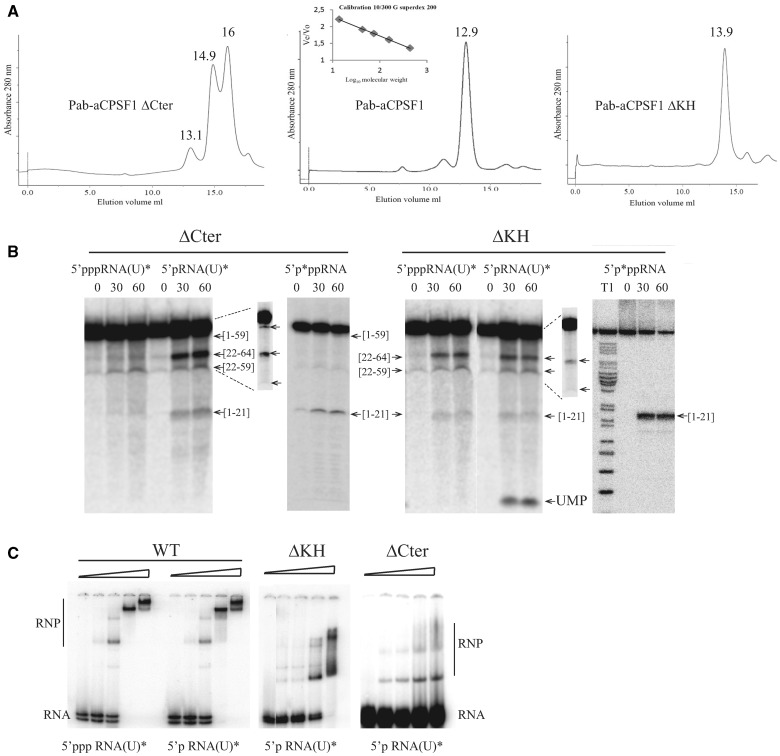
Figure 6.
Pab-aCPSF1ΔCter (ΔCter) and Pab-aCPSF1ΔKH (ΔKH) variants. (A) Size exclusion chromatography of Pab-aCPSF1 and its variants. The proteins were separated on Superdex 200 and the apparent molecular masses defined using the inset calibration curve. Gel filtration yielded a minor peak at 13.1 ml and major peaks at 14.9 and 16 ml for Pab-aCPSF1ΔCter corresponding to dimeric protein (148 kDa), monomers (55 kDa) and a proteolysis products (30 kDa) (Supplementary Figure S4), respectively. It should be noted that the monomer elutes at higher volume than expected (55 kDa instead of 72 kDa) (Supplementary Figure S4) (left). Gel filtration yielded a peak at 12.8 ml for Pab-aCPSF1 corresponding to a dimeric protein (159 kDa) (middle) Gel filtration yield a peak at 13.9 ml for Pab-aCPSF1ΔKH corresponding to dimeric protein (92 kDa) (right) (B) Kinetic analysis of RNA cleavage of 5′p*pp RNA, 5′ppp RNA(U)* and 5′p RNA(U)* substrates by protein variants (6 µM) at 65°C for the indicated times. Symbols are as in Figure 2. The products of the reaction were analysed on a 10% PAGE. Long migrations are shown in the inset (dotted lines) for 5′p RNA(U)* products. (C) Gel retardation assay with 10 fmol of 5′p RNA(U)* or of 5′ppp RNA(U)* incubated with increasing concentrations (0, 0.3, 0.6, 1.25 and 2.5 µM) of Pab-aCPSF1 (WT), ΔKH or ΔCter variants. RNA-protein complexes (RNP) are indicated.