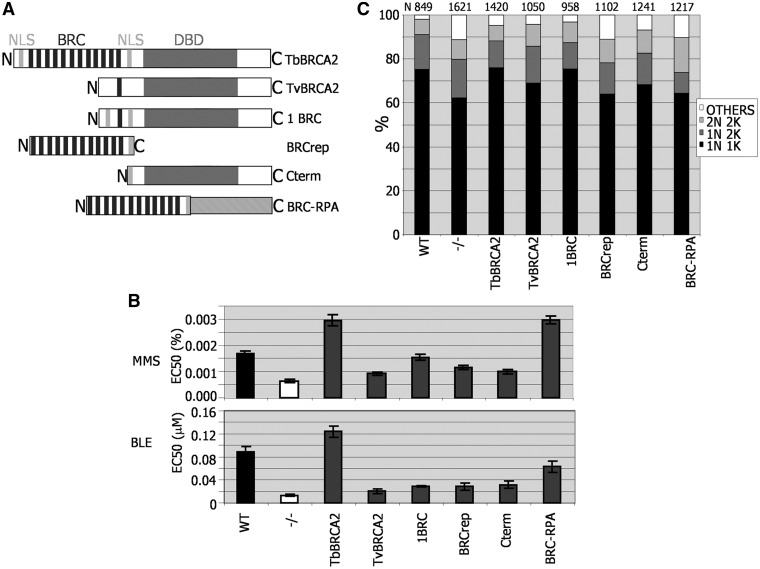
Figure 7.
Assessing T. brucei BRCA2 variant function in DNA repair and replication. (A) Diagrammatic representation of BRCA2 variants expressed in Lister427 BSF brca2−/− mutants, using the same strategy as described in Figure 3. The full-length T. brucei BRCA2 used here (TbBRCA2) contains 12 BRC repeats, a conserved DBD and two putative NLS. BRCA2 from T. vivax (TvBRCA2) has a single BRC repeat and a conserved DBD, but no NLSs have been predicted. 1BRC is identical to full-length T. brucei BRCA2, but the BRC array is reduced to a single repeat (Figure 3). BRCrep is a polypetide fragment of T. brucei BRCA2 encompassing only the BRC repeats and 33 downstream amino acids, including a bipartite NLS. Cterm is a 747 amino acid residue N-terminal truncation of T. brucei BRCA2. BRC–RPA is a fusion of the BRCrep polypeptide to the 50 kDa T. brucei replication protein A subunit. (B) The concentration of MMS or phleomycin (BLE) that caused 50% growth inhibition (EC50) of WT cells is compared with brca2−/− mutants, and with −/− cells expressing the BRCA2 variant polypeptides detailed in A. Values are the means from three experiments; bars indicate standard error. (C) The relative proportions of cells with standard (2N2K, 1N2K and 1N1K) or aberrant (other) nuclear (N) and kinetoplast (K) configurations are compared in populations of WT cells, brca2−/− mutants and −/− cells expressing the BRCA2 variant polypeptides detailed in A. N–K configurations were analysed by DAPI staining and microscopic examination; the number of cells counted (N) is indicated.