Figure 3.
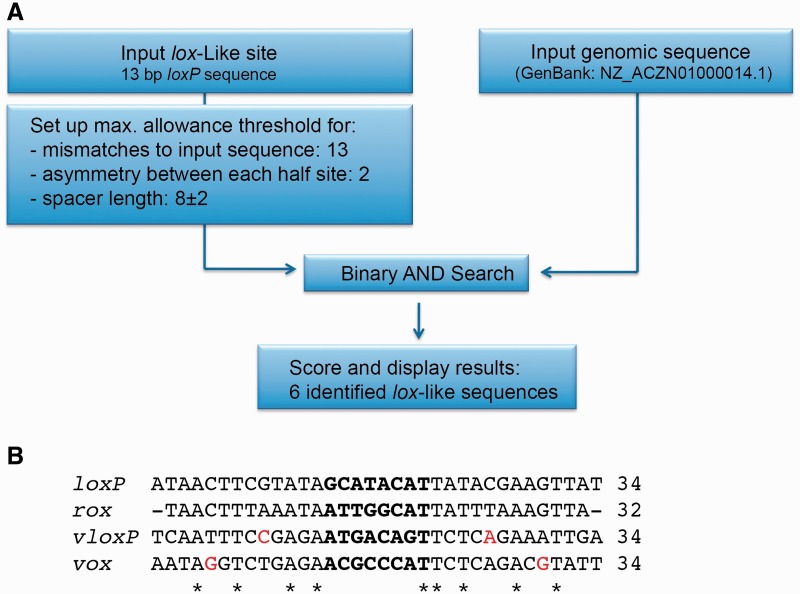
Identification and comparison of the vox target site. (A) Workflow of the SeLOX algorithm and important input parameters to predict lox-like sites in the genome of V. coralliilyticus strain NZ_ACZN01000014.1. The SeLOX results are shown in Supplementary Table S4. (B) Nucleotide alignment of vox with recombination sites for Cre (loxP), Dre (rox) and VCre (VloxP). Nucleotides that break perfect palindromic symmetry of the lox sites are colored in red. Asterisks depict conservation among all sites.