Abstract
In all living cells, protein synthesis occurs on ribonucleoprotein particles called ribosomes. Molecular models have been reported for complete bacterial 70S and eukaryotic 80S ribosomes; however, only molecular models of large 50S subunits have been reported for archaea. Here, we present a complete molecular model for the Pyrococcus furiosus 70S ribosome based on a 6.6 Å cryo-electron microscopy map. Moreover, we have determined cryo-electron microscopy reconstructions of the Euryarchaeota Methanococcus igneus and Thermococcus kodakaraensis 70S ribosomes and Crenarchaeota Staphylothermus marinus 50S subunit. Examination of these structures reveals a surprising promiscuous behavior of archaeal ribosomal proteins: We observe intersubunit promiscuity of S24e and L8e (L7ae), the latter binding to the head of the small subunit, analogous to S12e in eukaryotes. Moreover, L8e and L14e exhibit intrasubunit promiscuity, being present in two copies per archaeal 50S subunit, with the additional binding site of L14e analogous to the related eukaryotic r-protein L27e. Collectively, these findings suggest insights into the evolution of eukaryotic ribosomal proteins through increased copy number and binding site promiscuity.
INTRODUCTION
In all three domains of life, protein synthesis in the cell is performed by large macromolecular machines called ribosomes (1–3). In bacteria, such as Escherichia coli, the 70S ribosome is formed from a small 30S subunit, comprising one 16S ribosomal RNA (rRNA) and 21 ribosomal proteins (r-proteins), and a large 50S subunit composed of a 5S and 23S rRNA and 33 r-proteins (3). X-ray structures of bacterial ribosomal particles have revealed a complex network of interactions between the rRNAs and r-proteins (4–6). In comparison, eukaryotic 80S ribosomes are larger and more complex than bacterial ribosomes. For example, the yeast small 40S subunit contains one 18S rRNA and 33 r-proteins, whereas the large 60S subunit comprises 5S, 5.8S and 25S rRNAs together with 46 large subunit r-proteins (3). Crystal structures of the Saccharomyces cerevisiae 80S ribosome (7) and Tetrahymena thermophila 40S and 60S subunits (8,9) have determined the architecture of the additional rRNA expansion segments (ES) and variable regions (VR) as well as the localization of the eukaryotic-specific r-proteins.
Genomic studies indicate that archaeal ribosomes have an intermediate complexity compared with bacterial and eukaryotic ones (10–12). Although a 6.6 Å cryo-electron microscopy (EM) map of the archaeal 70S ribosome exists, no molecular model was reported (13). So far, the large 50S subunit from the Euryarchaeota Haloarcula marismortui has been crystallized disclosing structures for the 5S and 23S rRNA as well as 27 r-proteins, 12 of which are archaeal/eukaryotic-specific (14). Recently, a cryo-EM structure of the Euryarchaeota Methanobacterium thermoautotrophicus 50S subunit was determined, leading to the identification of five additional archaeal/eukaryotic specific r-proteins and some rRNA ES that are not present in the H. marismortui 50S subunit structure (15). Here, we present the complete molecular model for the Pyrococcus furiosus 70S ribosome, using a 6.6 Å cryo-EM structure (13). Based on 2D-PAGE and mass spectrometry (MS) analysis of Thermococcus kodakaraensis ribosome, coupled with additional cryo-EM reconstructions of Euryarchaeota Methanococcus igneus and T. kodakaraensis 70S ribosomes, and Crenarchaeota Staphylothermus marinus 50S subunit, we reveal a surprising promiscuity of r-proteins within archaeal ribosomes that has implications for the evolution of r-proteins in archaea and eukaryotes.
MATERIALS AND METHODS
Growth of M. igneus, S. marinus and T. kodakaraensis
T. kodakaraensis KOD1T (JCM 12380T, ATCC BAA-918T), M. igneus Kol5T (DSM 5666) and S. marinus F1T (DSM 3639, ATCC 49053T) were obtained from the culture collection of the Institute of Microbiology and Archaea Centre, University of Regensburg. T. kodakaraensis and S. marinus were grown under anoxic conditions in Marine-Thermococcus-medium (16) at 85°C and pH 7.0. Substrates 0.1% yeast extract and 0.1% peptone were added. For the cultivation of S. marinus, the medium was further supplemented with 0.7% elemental sulfur. The gas phases consisted of N2/CO2 (80/20 v/v, 250 kPa). M. igneus was grown in Methanotorris-medium at 85°C as previously described (17). The Methanotorris-medium consisted (per liter) of K2HPO4 0.0556 g, KH2PO4 0.0558 g, KCl 0.269 g, NaCl 25.14 g, NaHCO3 0.84 g, CaCl2 × 2H2O 0.368 g, MgCl2 × 6H2O 7.724 g, NH4Cl 1.180 g, Fe(NH4)2(SO4)2 × 6H2O 12 mg, Na3-Nitrilotriacetate (Titriplex I) 88 mg, Na2SeO4 8 mg, Na2WO4 × 2H2O 3.2 mg, Na2MoO4 × 2H2O 2.4 mg and 10 ml of trace element solution (18). The medium was covered with a gas phase of H2/CO2 (80/20 v/v, 250 kPa) and chemically reduced with Na2S x 9H2O (0.4 g/l). Mass cultivations for all strains were carried out in 300l enamel-protected fermenters (HTE, Bioengineering, Wald, Switzerland) with 250l culture medium and continuous supply of 0.1% sodium sulfide for pH stabilization. The cells were harvested by centrifugation (Padberg, Lahr, Germany), shock-frozen in liquid nitrogen and stored at −80°C until further use.
Preparation of ribosomal particles
Archaeal ribosomes were isolated and purified in a similar fashion as previously reported (19,20); however, with some modifications as described in (21): Cell pellets were dissolved in Tico buffer (20 mM Hepes pH 7.5, 10 mM Mg(OAc)2, 30 mM NH4OAc, 4 mM ß-Mercaptoethanol) at 4°C and subsequently disrupted by using a Microfluidizer (Microfluidics M-110L Pneumatic) at 18 000 psi. The crude homogenate was centrifuged twice at 30 000g at 4°C for 30 min to obtain the S30 fraction. A crude ribosomal fraction was obtained by centrifugation at 100 000g for 5 h at 4°C and dissolving the pellet in an equal volume of high salt wash (HSW) buffer (20 mM Hepes, 10 mM Mg(OAc)2, 500 mM NH4OAc, 4 mM ß-Mercaptoethanol, pH 7.5). Large debris were removed by centrifuging the crude ribosomes for 5 min at 18 000g at 4°C. The clear supernatant was diluted 10-fold in HSW buffer and layered on top of 1.3 volumes of 25% (w/v) sucrose cushion prepared in HSW buffer and centrifuged at 100 000g for 7 h at 4°C. The pellet was resuspended in a minimal volume of Tico buffer and subsequently purified using a sucrose-density gradient centrifugation (10–40% sucrose in Tico buffer) at 46 000g for 17 h at 4°C. Fractions corresponding to the 50S and 30S were separately pooled and pelleted at 140 000g for 12 h at 4°C and resuspended in a minimal volume of Tico buffer.
Extraction of the total ribosomal proteins, 2D-PAGE and MS
The total proteins from HSW ribosomes and purified ribosomal subunits were extracted by acetic acid according to Nierhaus et al. (22). Lyophilized proteins were further processed for liquid chromatography tandem MS (LC-MS/MS) analysis and for 2D-PAGE. Around 2 μg of total proteins was necessary for the LC-MS/MS, whereas 5–10 μg of total proteins were required for 2D-PAGE. The 2D electrophoresis was performed as described by Kaltschmidt and Wittman (23). LC-MS/MS analysis of 2D-gel spots on the LTQ ion-trap and of protein samples on the Orbitrap XL instrument was performed as previously described (21).
Electron microscopy
Cryo-EM and single particle reconstruction
As described previously (24), M. igneus and T. kodakaraensis 70S ribosomes and S. marinus 50S subunits were applied to carbon-coated holey grids. Images were collected on a Tecnai G2 Spirit TEM at 120 kV at a nominal magnification of 90 000 using a 2 K Eagle (2048 × 2048) CCD camera (FEI) resulting in a pixel size of 3.31 Å/pixel. The data were analyzed by determination of the contrast transfer function using CTFFIND (25), and further processed with the SPIDER software package (26), using the H. marismortui 50S subunit (PDB3CC2) (14) filtered to between 20–25 Å, as an initial reference. Further steps involved subsequent refinement and iterative sorting for heterogeneities. For the final reconstructions, 8932 particles were used for M. igneus, 10 431 for T. kodakaraensis and 11 142 for S. marinus; this resulted in 18 Å, 19 Å and 24 Å electron density maps at 0.5 FSC, respectively.
Modeling and figure generation
P. furiosus rRNA modeling
P. furiosus 16S and 23S/5S rRNA sequences were taken from GeneBank Accession number (Acc.) U20163 and Acc. AE009950, respectively. Structure-based sequence alignments of the conserved rRNA core were constructed using Sequence to Structure (S2S) (27) based on X-ray structures of the small ribosomal subunit of Thermus thermophilus for the 16S rRNA [Protein Data Bank (PDB) code 1fjf] (28) and the large ribosomal subunit of H. marismortui for the 23S and 5S rRNAs (PDB 1ffk) (14). For regions like the stalk base (H42–H44), H69, the L1 stalk (H76–H79) and the base of H98 (ES39L), the corresponding structures of the large ribosomal subunit of T. thermophilus and Escherichia coli (PDBs 2 × 9u and 3r8s) (29,30) were used as templates. Two eukaryote-like rRNA parts (h33 and H54–H59) were generated in separate S2S alignments based on the X-ray structure of the 80S ribosome from S. cerevisiae (PDBs 3o58 and 3o2z) (7). All remaining parts of the structure were built de novo using Assemble (31) essentially as described (32). The resulting complete models of the P. furiosus rRNAs were manually inspected and adjusted according to features of the electron density using Coot (33).
P. furiosus r-protein modeling
We used a 6.6 Å cryo-EM map of P. furious 70S ribosome (13) to localize and build models for 62 archaeal proteins (66 when including L8e(2), L8e(S), S24e(L) and L14e(2)). This consists of 33 r-proteins common to all three domains, 30 archaea/eukaryote-specific r-proteins (if LX is considered as L20e). A total of 27 proteins from the 50S subunit were modeled using archaeal X-ray H. marismortui protein templates (PDB 3cc2). In all, 25 r-proteins from the 30S subunit and five proteins from the 50S subunit (L14e, L34e, L35ae, L40e and L41e) were modeled using templates from the eukaryotic S. cerevisiae X-ray structure (PDBs 3u5g and 3u5c for the small subunit, PDBs 3u5e and 3u5i for the large subunit). Protein LX was modeled using Methanobacterium thermoautothropicus PDB 2jxt; the stalk protein P0 was based on Pyrococcus horikoshii and Methanocaldococcus janaschii, PDBs 3a1y and 3jsy, respectively; the L1 protein was based on T. thermophilus (Bacteria) PDB 2hw8 template. The multi-sequence alignment was performed using ClustalW (34), whereas for sequence analysis, Jalview was used. Protein models were created using Modeller (35) and further fit and analyzed using Chimera (36) (rigid body fitting) and Coot (33) (manual adjustments), as well as MDFF (37) in VMD (38).
Refinement and fitting of the rRNA and r-proteins into the EM densities
MDFF was used to refine the proteins and RNA into the density while fixing protein–RNA and protein–protein clashes, followed by an MDFF refinement of the entire 70S model.
Figure generation
Figures showing electron densities and atomic models were generated using Chimera (36).
RESULTS
A cryo-EM map and model for the archaeal 70S ribosome
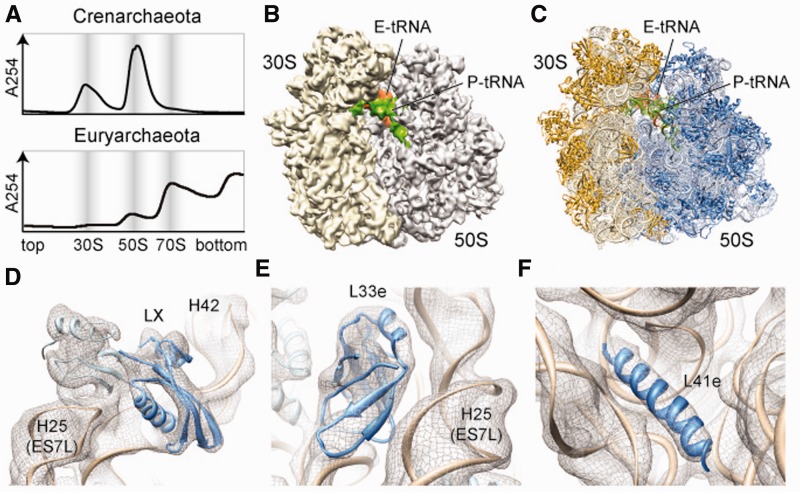
In contrast to purifications of archaeal ribosomal particles from Crenarchaeota Sulfolobus acidocaldarius, Pyrobaculum aerophilum (19,21), S. marinus (Figure 1A, upper panel) and Euryarchaeota M. thermoautotrophicus (15) where only 30S and 50S subunits were obtained, we were able to isolate intact archaeal 70S ribosomes from Euryarchaeota P. furiosus and T. kodakaraensis translation extracts using sucrose gradient centrifugation (Figure 1A, lower panel). A cryo-EM map of the P. furiosus 70S ribosome at 6.6 Å (0.5 FSC) (Figure 1B) (13) was then used to generate a molecular model for the rRNA and r-protein components (Figure 1C). The P. furiosus rRNAs were built in S2S (27) and Assemble (31), using initial models based on templates derived from the X-ray structures of the bacterial 30S (28) and 70S (29,30), the archaeal 50S (14) and the eukaryotic 80S (7) (see Materials and methods for details). Thus, in addition to the conserved rRNA core, five variable regions [VR5S (h16-h17), VR8S (h33), VR1L (H1), VR22L (H58) and H16-18] and 12 ES (ES7S, ES9S, ES4L, ES5L, ES7L, ES9L, ES15L, ES20L, ES24L, ES26L, ES39L and ES41L) were modeled (Supplementary Figure S1). The majority of the P. furiosus VRs and ESs had conformations remarkably similar to the equivalent regions in the eukaryotic ribosome (7–9,32) (Supplementary Figure S2), as noted previously for the M. thermoautotrophicus 50S subunit (15). However, VR5S (h16), VR1L (H1) and ES39L adopt novel conformations in the P. furiosus 70S (Supplementary Figure S3) that have not been observed in previous ribosome structures. Unlike the M. thermoautotrophicus 23S rRNA, which is a composite of H. marismortui and T. thermophila rRNA fragments (15), we present complete models for the P. furiosus 16S and 23S rRNAs (Supplementary Figure S4) with continuous P. furiosus sequence and numbering (Supplementary Figures S1, S5–S7).
Figure 1.
Cryo-EM structure and molecular model of an archaeal 70S ribosome. (A) Sucrose density gradient centrifugation profile of ribosomal particles from the Crenarcheaota S. marinus (Upper panel) and Euryarchaeota T. kodakaraensis (lower panel), with 30S, 50S and 70S peaks highlighted. (B) Cryo-EM reconstruction (30S, yellow; 50S, gray) and (C) molecular model (16S and 23 rRNA, light yellow and blue; SSU and LSU r-proteins, gold and blue) of the P. furiosus 70S ribosome. P- and E-tRNA are colored green and orange, respectively. (D–F) Fit of molecular models for rRNA (tan) and r-proteins (blue), (D) LX, (E) L33e and (F) L41e, into the cryo-EM density of the P. furiosus 70S ribosome (gray mesh).
Based on genomic analysis, the P. furiosus 70S ribosome is predicted to contain 64 r-proteins, 25 in the 30S and 39 in the 50S subunit (10–12) (Supplementary Tables S1–S4). In all, 35 (30S, 15; 50S, 20) of the P. furiosus r-proteins have counterparts in bacteria, and the location of an additional 12 large subunit r-proteins is known from the X-ray structure of the H. marismortui 50S subunit (14). Locations for the remaining 17 (30S, 10; 50S, 7) were determined by homology with the respective eukaryotic r-proteins present in the X-ray structure of the S. cerevisiae 80S ribosome (7). The models for P. furiosus r-proteins L14e [we use the revised and simplified nomenclature based on family names for eukaryotic r-proteins (7), see Supplementary Tables S1–S4], L30e, L34e, L40e and LX (Figure 1D) were in agreement with those reported recently for the euryarchaeotal M. thermoautotrophicus 50S subunit (15). In addition to 10 models of small subunit P. furiosus r-proteins, we also present models for r-proteins L33e (Figure 1E) and L41e (Figure 1F), which are absent in the genomes of H. marismortui and M. thermoautotrophicus (Supplementary Tables S1 and S2) (10–12). The high quality of the P. furiosus 70S ribosome cryo-EM map enabled an accurate fit of the molecular models of the rRNA and r-proteins by using distinct features of the electron density seen for the major and minor grooves of the RNA helices and rod-like densities for r-proteins (Supplementary Figure S4). Surprisingly, after fitting all rRNA and r-protein, four regions of additional density remained unaccounted for; one located on the 30S subunit and three on the 50S subunit. As all the rRNA was satisfactorily modeled and the additional density had features reminiscent of protein, we hypothesized that the additional density was owing to the presence of yet unidentified r-proteins.
2D-PAGE and MS of Euryarchaeota ribosomes
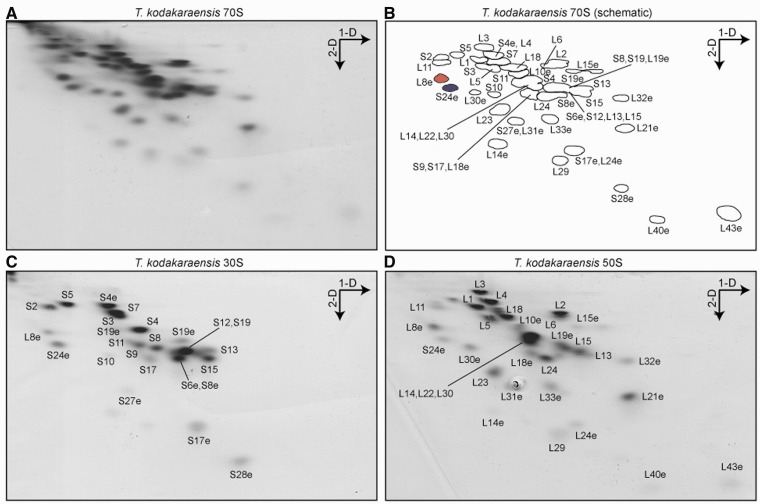
Our previous MS analysis of the Crenarchaeota S. acidocaldarius and P. aerophilum ribosomes led to the identification of a number of hypothetical proteins with basic isoelectric point that were ribosome associated (21). Thus, to search for additional r-proteins in Euryarchaeota, we performed 2D-PAGE (23) on high-salt washed 70S ribosomes from T. kodakaraensis (previously called Pyrococcus kodakaraensis) (Figure 2A and B), which belongs to the same Thermococcaceae family as P. furiosus. MS identification of the protein spots, coupled with LC-MS/MS analysis of the total protein samples, led to the identification of 25/25 (100%) and 36/38 (95%) of the 30S and 50S subunit r-proteins, respectively (Supplementary Tables S3 and S4). Additionally, the T. kodakaraensis 70S ribosomes were split into 30S and 50S subunits, isolated using sucrose gradients and also analyzed by 2D-PAGE and MS (Figure 2C and D). Surprisingly, MS revealed that a protein spot for large subunit r-protein L8e (L7ae) was also present in the 2D-PAGE of the 30S subunit, which, although relatively weak, had similar intensity to some bona fide small subunit r-proteins, such as S10, S17 and S19e (Figure 2C). Similarly, MS identified a protein spot for the small subunit r-protein S24e in the 2D-PAGE of the 50S subunit, with comparable intensity to large subunit r-proteins L11, L16 and L35 (Figure 2D).
Figure 2.
2D-PAGE and MS analysis of Euryarchaeota ribosomal proteins. (A–D) Coomassie blue-stained 2D gel of (A) high-salt-washed T. kodakaraensis 70S ribosomes (MS assignments of protein spots labeled in (B) scheme), and sucrose gradient purified (C) 30S and (D) 50S subunits. The direction of the first (1D, based on charge) and second (2D, based on mass) dimensions of electrophoresis are indicated with arrows, and spots for L8e (L7ae) and S24e are colored.
Identification and localization of promiscuous archaeal r-protein L8e (L7ae)
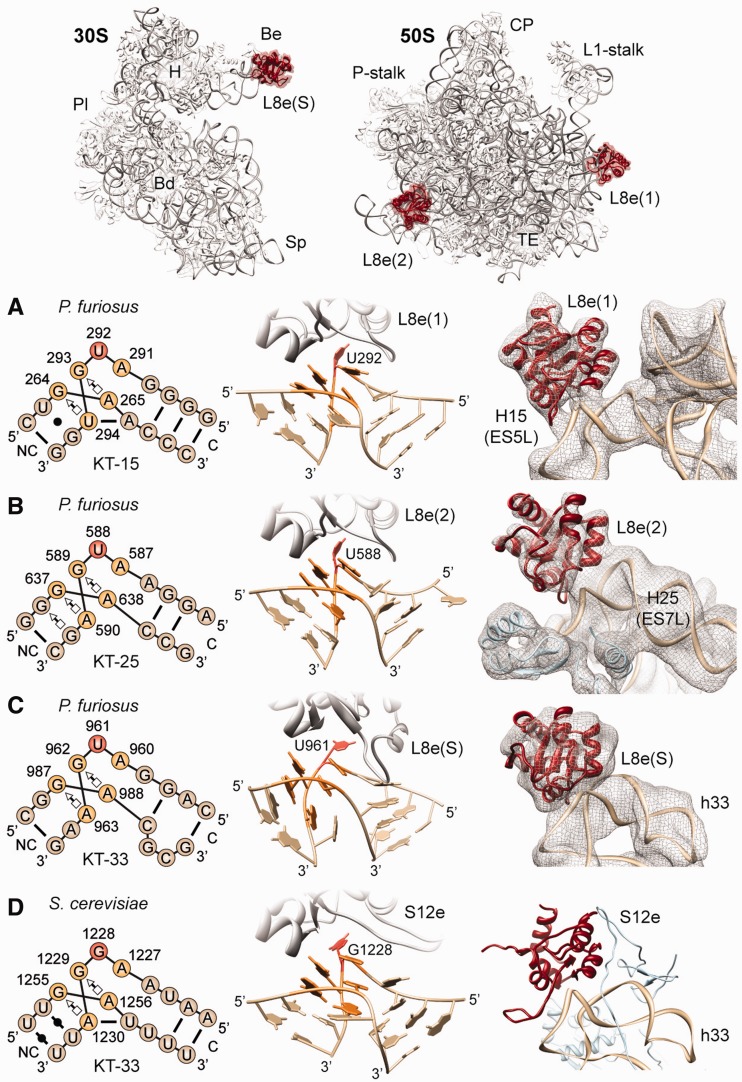
L8e (L7ae) is a compact globular protein with a fold consisting of alternating α-helices and β-strands, forming an α-β-α sandwich structure that interacts with a kink-turn (KT) motif (39,40)—an asymmetric internal loop that induces a sharp bend in the phoshodiester backbone of an RNA helix (39). In the archaeal and eukaryotic large subunit structures (7,9,14), L8e interacts with the KT motif present in helix 15 of the 23S rRNA (termed KT-15) by specifically recognizing a bulged uridine nucleotide (U292) located within the internal loop (Figure 3A). Thus, to ascertain whether L8e has an additional binding site on the small and/or large subunit, we searched for similar KT motifs within the 16S and 23S rRNAs of the T. kodakaraensis and P. furiosus 70S ribosome. Two further KT motifs with bulged uridine nucleotides were identified in direct vicinity of the unassigned densities in the P. furiosus 70S cryo-EM map, one in H25 of the 23S rRNA (KT-25, Figure 3B) and the other in h33 of the 16S rRNA (KT-33, Figure 3C). Docking of the model for L8e into each of the unassigned densities yielded an excellent fit and maintained canonical interaction with the bulged uridine nucleotide of the respective KT motifs (Figure 3B and C). Together with the MS data, these findings suggest that L8e has three binding sites in the P. furiosus 70S ribosome, namely, the canonical site, L8e(1), positioned adjacent to the L1 stalk; a second site, L8e(2), interacting with H25 (ES7L) at the back of the large subunit; as well as a third site, L8e(S), located on the beak of the small subunit. This promiscuity of L8e already has a precedent, as L8e (L7ae) is known to also interact with bulged uridine nucleotides found within KT motifs of the archaeal C/D and H/ACA box archaeal small nucleolar ribonucleoprotein particle (snoRNP) RNAs (Supplementary Figure S8) (41,42) as well as within the KT-containing archaeal RNase P RNA (43).
Figure 3.
Promiscuity of archaeal ribosomal protein L8e (L7ae). (A–D) Schematic (left) and structural (middle) representations of KT motif (A) KT-15 of the canonical L8e(1) binding site and (B) KT-25 at the L8e2 position, on the 50S subunit, compared with KT-33 at the (C) L8e(S) binding site on the P. furiosus 30S subunit and (D) S12e binding site on the S. cerevisiae 40S subunit (7). Right-hand panels show a fit of molecular models to the cryo-EM density (mesh) of P. furiosus 70S ribosome for (A) L8e(1) and (B) L8e(2) on the 50S subunit, and (C) L8e(S) on the small subunit, and in comparison (D), the binding position of S12e on the S. cerevisiae 40S subunit (7). Insets at top of figure show the overview of the L8e-binding positions (red) on the small (left) and large (right) ribosomal subunit. Major landmarks are indicated: beak (Be), body (Bd), platform (Pl), head (H), spur (Sp), central protuberance (CP) and tunnel exit (TE). C and NC indicate the canonical and non-canonical stem in KT diagrams. The cross-correlation of the fit of L8e crystal structure to the density for the different binding sites is as follows: L8e(1) = 0.90; L8e(2) = 0.87; L8e(S) = 0.81.
In eukaryotic 60S subunits, ES7L is elongated and KT-25 is absent, thus the L8e(2) binding site is not present (7,32,44) (Supplementary Figure S9). In contrast, KT-33 is present in all eukaryotic 40S subunits, where it forms part of the binding site for eukaryotic-specific r-protein S12e (Figure 3D)—a protein with the same fold as L8e (Supplementary Figure S10). In fact, S12e actually belongs to the evolutionary conserved L8e (L7ae) family of KT-binding proteins, which also encompasses r-protein L30e as well as the dual spliceosome/snoRNP 15.5kD protein (Snu13p in yeast), RNase P component Rpp38 (Pop3p), the snoRNP protein NHP2 and SBP2, a protein that binds the selenocysteine insertion sequence element RNA (39,45,46). However, S12e recognizes a bulged-out guanine (G1228) nucleotide (rather than a uridine as L8e) within the internal loop of KT-33 (Figure 3D), which is conserved in eukaryotic 18S rRNAs (Supplementary Figure S11). Substitution of uridine to guanine in KT-containing RNAs reduces binding affinity of archaeal L8e by ∼100-fold (47), indicating how eukaryotic 80S ribosomes ensure specificity of L8e and S12e binding to KT-15 and KT-33, respectively.
Identification and localization of promiscuous archaeal r-proteins S24e and L14e
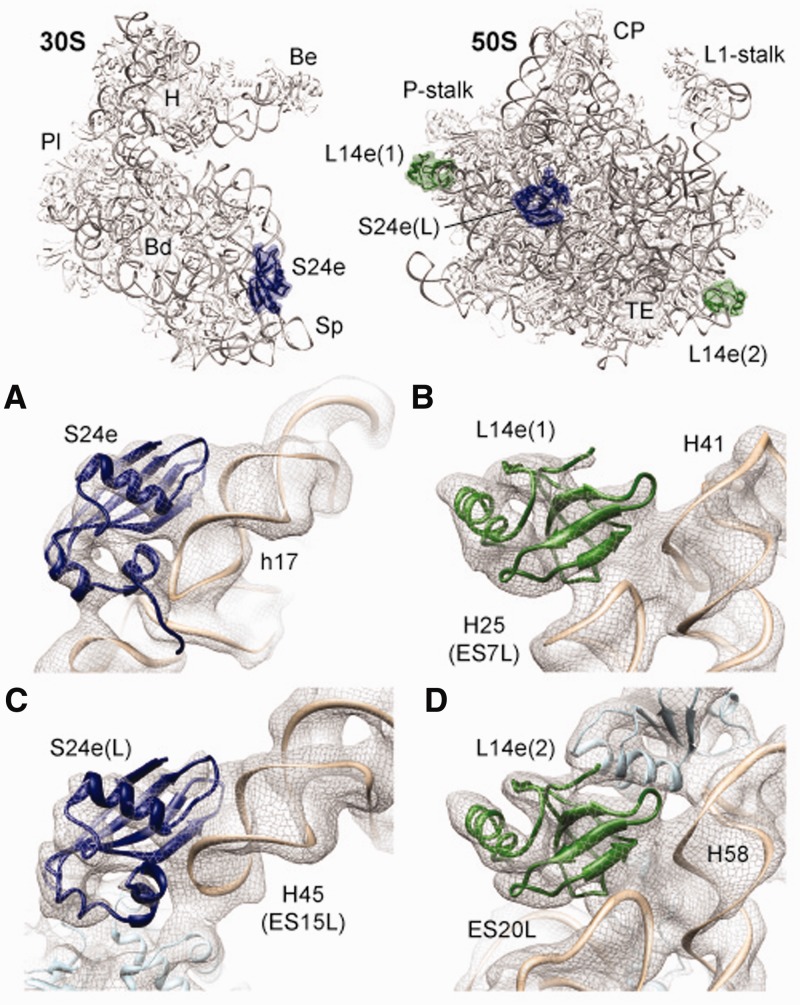
Based on the 2D-PAGE and MS data, we examined whether r-protein S24e also has a binding site on the 50S subunit that could account for one of the remaining unassigned densities in the P. furiosus 70S cryo-EM map. S24e comprises a four-stranded anti-parallel β-sheet flanked by three short α-helices and contains an RNA recognition motif, similar to the related r-protein L23 (48). The canonical binding site for S24e is located at the base of the body of the small subunit, where the terminal β-strand of S24e interacts with the major groove of h17 (Figure 4A). We found an excellent fit of the model for S24e to one of the unassigned densities on the large subunit, similarly oriented such that the terminal β-strand of S24e interacts with major groove of an RNA helix, in this case H45 (ES15L) (Figure 4B). ES15L is slightly longer and adopts a different conformation in eukaryotic 80S ribosomes, consistent with the absence of a second S24e(2)-binding position (7,9,32) (Supplementary Figure S9).
Figure 4.
Promiscuity of archaeal ribosomal proteins S24e and L14e. (A–D) Fit of molecular models of (A-B) S24e (blue) and (C-D) L14e (green) into the cryo-EM density (gray mesh) of P. furiosus 70S ribosome, namely, (A) S24e on the 30S subunit and (B) S24e(S) on the 50S subunit, and (C) L14e(1) and (D) L14e(2) on the 50S subunit. Insets at the top show the overview of the S24e (blue) and L14e (green)-binding positions on the small (left) and large (right) ribosomal subunit. Major landmarks are indicated as in Figure 3.
Given the surprising finding that archaeal r-proteins, such as L8e and S24e, were present in more than one copy per P. furiosus 70S ribosome, we next examined whether the last unassigned density could be also attributed to another promiscuous r-protein. Indeed, we found that L14e could be unambiguously fit to the remaining electron density located on the 50S subunit (Figure 4C). Archaeal L14e has an Src-homology 3 (SH3)-like β-barrel (49) and in the canonical position on the ribosome, L14e(1), is located at the back of the central protuberance adjacent to LX, where it interacts with the backbone of H41 and the tip of H25 (ES7L) (Figure 4C) (7,9,15,44). Analogously, the second L14e(2) interacts with the backbone of H58 and the tip of ES20L (Figure 4D). In eukaryotic 80S ribosomes, the binding position of L14e(2) is occupied by the related eukaryotic-specific r-protein L27e, which also contains an SH3-like β-barrel (7,9,14,15) (Supplementary Figure S9).
Taxonomic distribution of the promiscuous archaeal ribosomal proteins
The discovery of inter- and intra-subunit promiscuity and multi-copy r-proteins within the P. furiosus 70S ribosome raised the question as to whether this represents a general phenomenon occurring in other archaeal species or whether it is specific for the Thermococcaceae family. To address this, we searched for the presence of additional binding sites of L8e, L14e and S24e in the available archaeal ribosomal structures, namely the 50S subunit from the Euryarchaeota H. marismortui (14) and M. thermoautotrophicus (15) as well as the Crenarchaeota S. acidocaldarius and P. aerophilum (21). In addition, we determined additional cryo-EM structures of the Euryarchaeota M. igneus and T. kodakaraensis 70S ribosomes at 18 Å and 25 Å (0.5 FSC), respectively, as well as the Crenarchaeota S. marinus 50S subunit at 24 Å (0.5 FSC) resolution. With the exception of the cryo-EM structure of the T. kodakaraensis 70S ribosome, we did not observe additional density in any of the cryo-EM maps for the presence of S24e on the large subunit (Supplementary Figure S12A–F), suggesting that S24e(L) is specific for the Thermococcaceae family. Consistently, S24e(L) was also not observed in the X-ray structure of the H. marismortui 50S subunit (14). In contrast, we observed additional density for a second binding position of L14e in all the cryo-EM maps (Supplementary Figure S12G–L), suggesting that L14e(2) is ubiquitous across the archaeal phylogeny. However, L14e(2) was not found in the X-ray structure of the H. marismortui 50S subunit (14), consistent with the finding that L14e has been lost in Halobacteria (10–12).
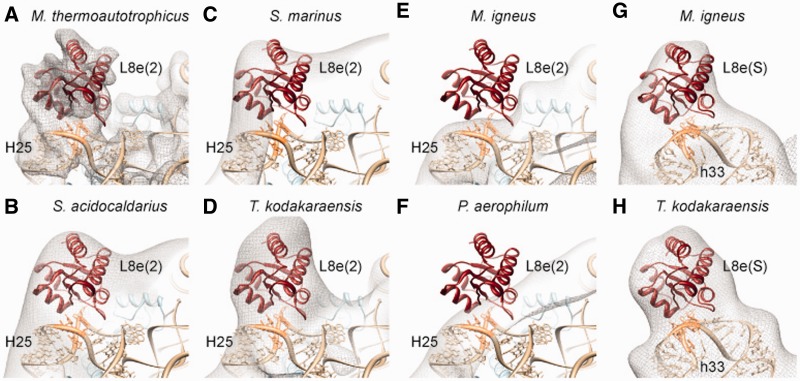
The presence of additional binding sites of L8e on archaeal ribosomes correlates perfectly with the expectations based on the presence or absence of the relevant KT motif in h33 (KT-33) and H25 (KT-25) of the 16S and 23S rRNA, respectively. Specifically, additional density was observed for L8e(2) on the 50S subunits of M. thermoautotrophicus, S. acidocaldarius, S. marinus and T. kodakaraensis (Figure 5A–D), all of which are predicted to contain KT motifs with a conserved uridine in the internal loop (Supplementary Figure S13), whereas no density was observed for L8e(2) in the cryo-EM maps of M. igneus and P. aerophilum 50S subunits (Figure 5E–F), where the KT motif has been lost (Supplementary Figure S13). Moreover, the KT motif is also absent in the X-ray structure of the H. marismortui 50S subunit, which lacks L8e(2) (14). In contrast, KT-33 with a conserved uridine in the internal loop is predicted for all archaeal 16S rRNA sequences (Supplementary Figure S11). Consistently, we observed additional density for L8e(S) on the small subunit in our two newly determined cryo-EM maps of the Euryarchaeota 70S ribosomes from M. igneus and T. kodakaraensis (Figure 5G–H). Moreover, previous 2D-PAGE and MS analysis of 30S subunits from the Crenarchaeota S. acidocaldarius also detected a protein spot for L8e (L7ae) with a stoichiometry similar to some small subunit r-proteins (21). Collectively, these findings lead us to suggest that L8e(S) is present in all archaeal ribosomes and should be considered as a bona fide archaeal small subunit r-protein.
Figure 5.
Distribution of additional L8e-binding sites on archaeal ribosomes. (A–H) The cryo-EM density (gray mesh) and fitted model for the P. furiosus 70S ribosome showing the presence of L8e(2) (red) in (A) M. thermoautotrophicus 50S (EMD-2012) (15), (B) S. acidocaldarius 50S (EMD-1797) (21), (C) S. marinus 50S, (D) 50S subunit of T. kodakaraensis 70S and absence in (E) 50S subunit of M. igneus 70S, (F) P. aerophilum 50S (EMD-1796) (21), as well as presence of L8e(S) in the (G) 30S of M. igneus 70S and (H) 30S of T. kodakaraensis 70S. Ribosomal RNA is shown in tan with KT motifs in orange.
DISCUSSION
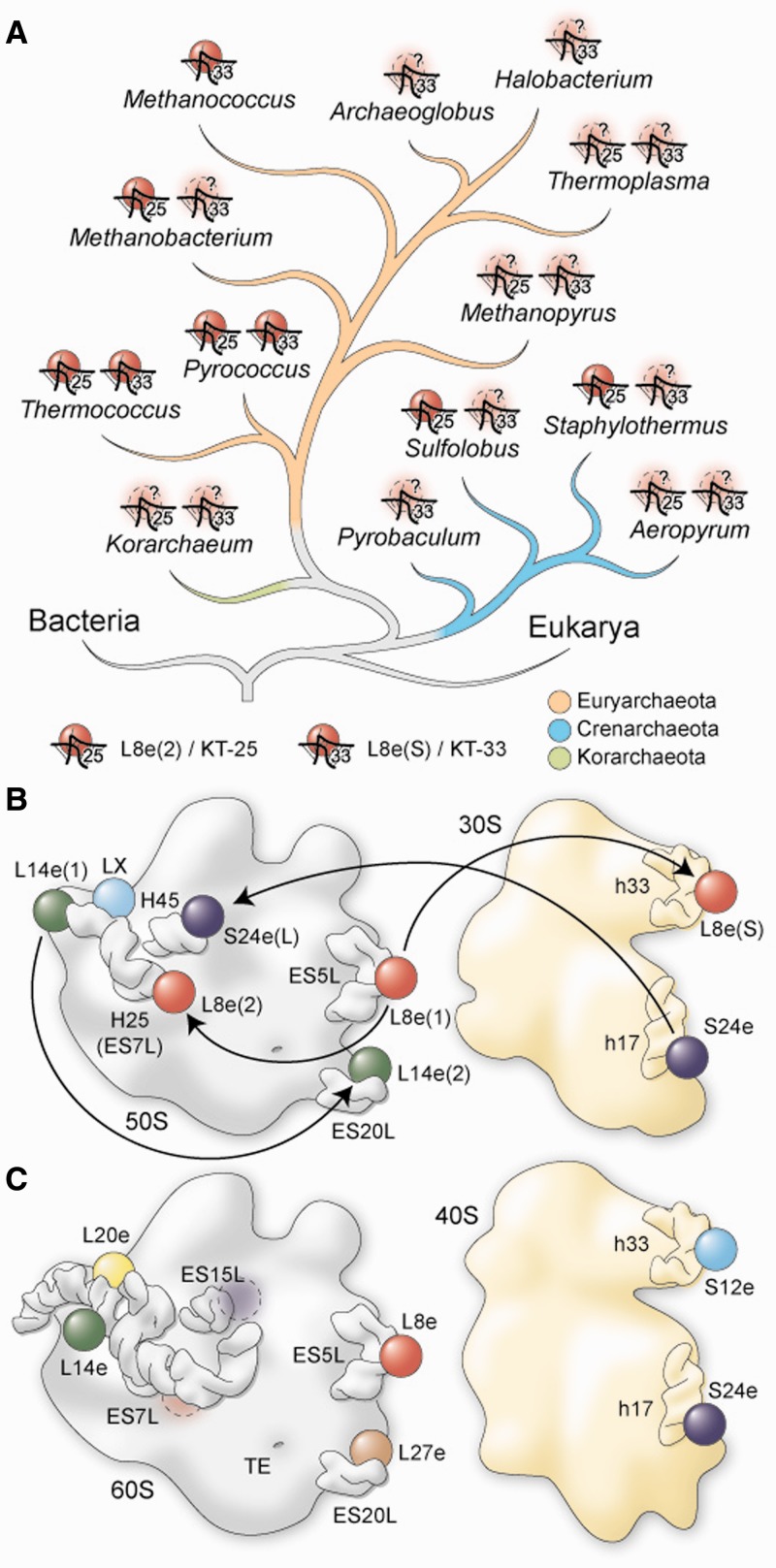
With the exception of the stalk proteins (L7/L12 in bacteria and P1-P3 in eukaryotes), r-proteins are thought to be present in one copy per ribosome. Here, we demonstrate that this concept does not hold true for archaeal ribosomes by showing that P. furiosus and T. kodakaraensis 70S ribosomes have two copies each of S24e and L14e as well as three copies of L8e. Moreover, based on our analysis of KT motifs across the complete archaeal phylogeny (Figure 6A), we predict that all archaeal ribosomes contain at least two copies of L8e, one at the canonical site on the 50S subunit and an additional site located on the small subunit (KT-33), whereas the second L8e binding site on the 50S subunit (KT-25) appears to be lost predominantly in late branching Euryarchaeota, such as Methanococcus, Archaeoglobus and Halobacterium. To our knowledge, the inter-subunit and intra-subunit promiscuity exhibited by S24e/L8e and L14e/L8e, respectively, has not been observed previously (Figure 6B). Furthermore, we find that the intersubunit promiscuity of S24e is specific for the Thermococcus/Pyrococcus 70S ribosomes, whereas the intrasubunit promiscuity of L14e appears to be widely established in archaea. The observation that L8e(S) and L14e(2) occupy the same binding position on the archaeal ribosome, as the related r-proteins S12e and L27e occupy on the eukaryotic ribosome (Figure 6B and C and Supplementary Figure S9), may reflect that S12e and L27e originally evolved from the promiscuous behavior of L8e and L14e, as has been suggested for archaeal LX and the related eukaryotic r-protein L20e (15) (Figure 6B and C and Supplementary Figure S9). Such a scenario would be consistent with the intermediate complexity of archaeal ribosomes compared with bacterial and eukaryotic ribosomes (Supplementary Figure S14) reflecting the potential for archaeal ribosomes to represent intermediate steps in the evolution of eukaryotic ribosomes.
Figure 6.
Inter- and intra-subunit promiscuity of archaeal ribosomal proteins. (A) Taxonomy of archaea, with presence or absence of KT-25, KT-33 and L8e(2)- or L8e(S)-binding sites. Dashed protein symbols indicate that their presence is predicted, but not experimentally proven yet. Phylogenetic tree is based on (50). (B) Schematic showing promiscuity of archaeal ribosomal proteins, compared with (C) the equivalent situation in the eukaryotic yeast 80S ribosome.
ACCESSION NUMBERS
The cryo-EM maps of the M. igneus 70S, T. kodakaraensis 70S and S. marinus 50S subunit have been deposited in the EM databank under accession numbers EMD-2172, EMD-2170 and EMD-2171, respectively, and the atomic models for the P. furiosus 30S and 50S subunits have been deposited in the protein databank under accession numbers PDB3j20 and 3j21/3j2L, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–15 and Supplementary references [51–53].
FUNDING
Deutsche Forschungsgemeinschaft [SFB594, SFB646 to R.B. and FOR1805 to R.B. and D.N.W.]; Fonds der chemischen Industrie (to S.F.); EMBO young investigator program (to D.N.W.). Funding for open access charge: LMU.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN, Cate JH. The structure and function of the eukaryotic ribosome. Cold. Spring. Harb. Perspect. Biol. 2012;4:1–17. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 5.Schuwirth B, Borovinskaya M, Hau C, Zhang W, Vila-Sanjurjo A, Holton J, Cate J. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Selmer M, Dunham C, Murphy FT, Weixlbaumer A, Petry S, Kelley A, Weir J, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 8.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 9.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 10.Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S. On the last common ancestor and early evolution of eukaryotes: reconstructing the history of mitochondrial ribosomes. Res. Microbiol. 2010;162:53–70. doi: 10.1016/j.resmic.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 15.Greber BJ, Boehringer D, Godinic-Mikulcic V, Crnkovic A, Ibba M, Weygand-Durasevic I, Ban N. Cryo-EM structure of the archaeal 50S ribosomal subunit in complex with initiation factor 6 and implications for ribosome evolution. J. Mol. Biol. 2012;418:145–160. doi: 10.1016/j.jmb.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller M, Braun FJ, Dirmeier R, Hafenbradl D, Burggraf S, Rachel R, Stetter KO. Thermococcus alcaliphilus sp. nov., a new hyperthermophilic archaeum growing on polysulfide at alkaline pH. Arch. Microbiol. 1995;164:390–395. [PubMed] [Google Scholar]
- 17.Burggraf S, Fricke H, Neuner A, Kristjansson J, Rouvier P, Mandelco L, Woese CR, Stetter KO. Methanococcus igneus sp. nov., a novel hyperthermophilic methanogen from a shallow submarine hydrothermal system. Syst. Appl. Microbiol. 1990;13:263–269. doi: 10.1016/s0723-2020(11)80197-9. [DOI] [PubMed] [Google Scholar]
- 18.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Londei P, Altamura S, Cammarano P, Petrucci L. Differential features of ribosomes and of poly(U)-programmed cell-free systems derived from sulfur-dependent archaebacterial species. Eur. J. Biochem. 1986;157:455–462. doi: 10.1111/j.1432-1033.1986.tb09689.x. [DOI] [PubMed] [Google Scholar]
- 20.Bommer U, Burkhardt N, Jünemann R, Spahn CMT, Triana-Alonso FJ, Nierhaus KH. In: Subcellular Fractionation. A Practical Approach. Graham J, Rickwoods D, editors. Oxford: IRL Press at Oxford University Press; 1996. pp. 271–301. [Google Scholar]
- 21.Marquez V, Frohlich T, Armache JP, Sohmen D, Donhofer A, Mikolajka A, Berninghausen O, Thomm M, Beckmann R, Arnold GJ, et al. Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J. Mol. Biol. 2011;405:1215–1232. doi: 10.1016/j.jmb.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 22.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from E. coli. Proc. Natl Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaltschmidt E, Wittmann HG. Ribosomal proteins. VII 2D polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal. Biochem. 1970;36:401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- 24.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA-anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 25.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 26.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 27.Jossinet F, Westhof E. Sequence to structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 28.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 29.Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc. Natl Acad. Sci. USA. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl Acad. Sci. USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD—Visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein D, Moore P, Steitz T. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 41.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 43.Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl Acad. Sci. USA. 2010;107:14573–14578. doi: 10.1073/pnas.1005556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis PP, Omer A. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 2005;8:685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Dlakic M. 3D models of yeast RNase P/MRP proteins Rpp1p and Pop3p. RNA. 2005;11:123–127. doi: 10.1261/rna.7128905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn JF, Tran EJ, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum. Mol. Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 49.Edmondson SP, Turri J, Smith K, Clark A, Shriver JW. Structure, stability, and flexibility of ribosomal protein L14e from Sulfolobus solfataricus. Biochemistry. 2009;48:5553–5562. doi: 10.1021/bi9003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr. Opin. Microbiol. 2011;14:274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue S, Wang R, Yang F, Terns RM, Terns MP, Zhang X, Maxwell ES, Li H. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol. Cell. 2010;39:939–949. doi: 10.1016/j.molcel.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.