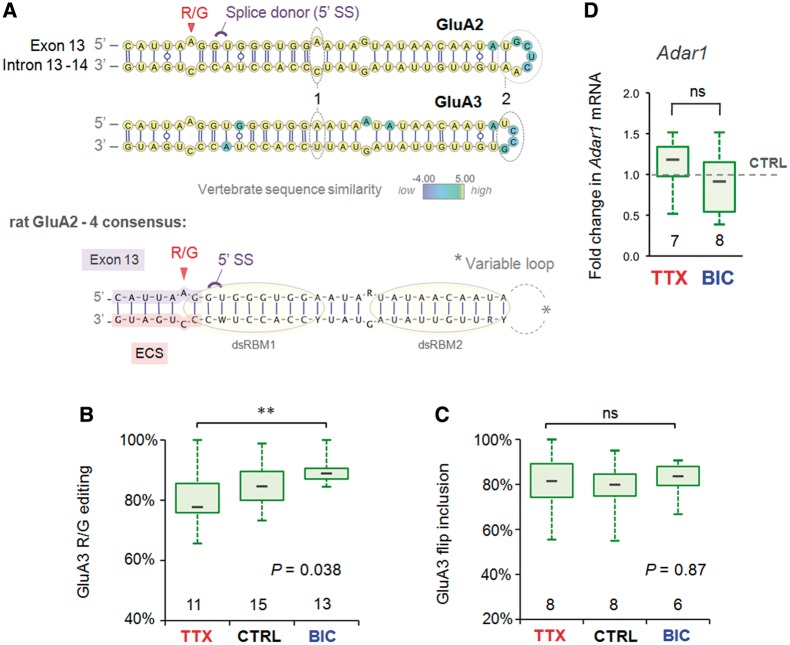
Figure 4.
Comparison of activity-dependent R/G site regulation and substrate properties between AMPA subunit paralogs. (A) Top: the predicted common secondary structure (γ-centroid estimator) of the GluA2 and 3 pre-mRNA encompassing the R/G site for 10 vertebrate species (Supplementary Table S1 and Supplementary Figure S6) mapped onto the rat sequences. Base pairs are rendered according to the Leontis/Westhof (LW) nomenclature. The splice donor site (5′ SS) and editing sites are annotated in violet and red, respectively. Base positions are color-coded according to vertebrate sequence similarity (see color map), thus yellow base positions are completely conserved. Note the mismatch (1) and the longer loop sequence (2) distinguishing GluA2 from the GluA3 R/G editing substrate. Bottom: the consensus structure of the three rat paralogs of the GluA2–4 R/G editing substrates. Lilac highlight = exon sequence; salmon highlight = R/G site ECS. The putative binding sites of the two dsRBMs of Adar2 are illustrated with yellow circles. (B) Changes in R/G editing of GluA3 show regulation by an activity similar to GluA2, albeit to a lesser extent. The number at the base of each plot is the number of slices. The P-value is derived from the Kruskal–Wallis ANOVA test statistic and the asterices summarize the results of Dunn’s post-tests: **P < 0.01. (C) The abundance of GluA3 flip splice variant as a fraction of total subunit mRNA expressed in CA1 is plotted for the different drug treatments. Quantification of splice variants is determined from mean peak height ratios for the first alternatively spliced nucleotide positions. GluA3 flip/flop splicing is invariant across drug treatments. (D) The fold change in CA1 mRNA levels of Adar1 relative to the mean of intra-PCR run CTRL treatments is plotted (box and whisker) for the (48 h) drug treatments. Adar1 expression was normalized to the endogenous house keeping gene Gapdh. Adar1 expression was comparable between CTRL and TTX. The two-tailed P-value and asterisks are derived from the Mann–Whitney U-test statistic, P = ns.