Fig 1.
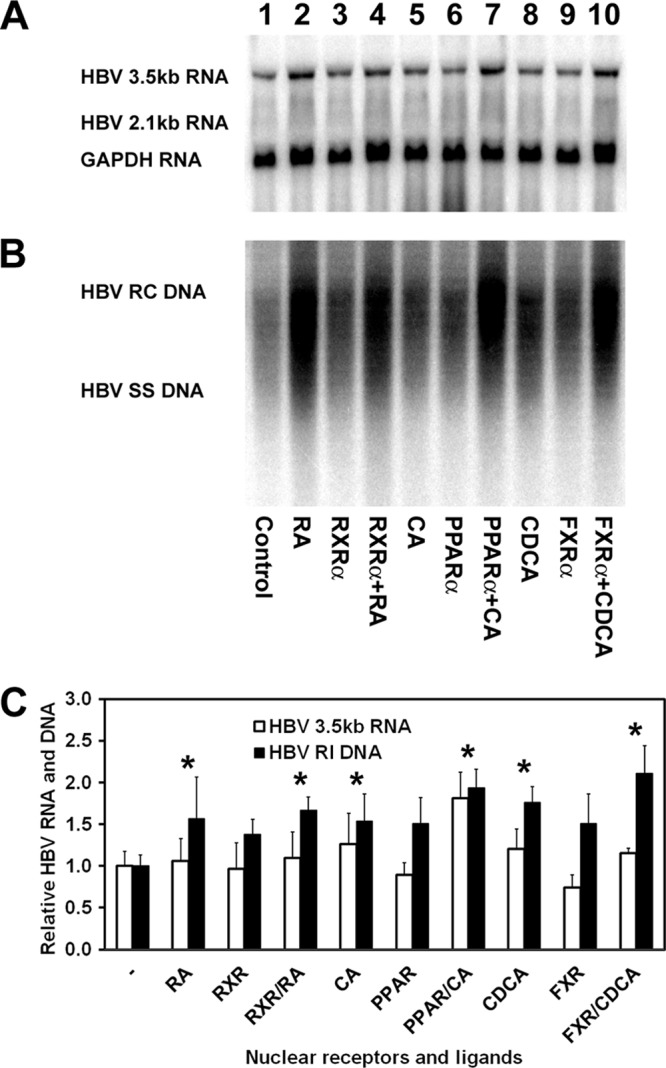
Effects of retinoids, peroxisome proliferators, and bile acids on HBV biosynthesis in the human hepatoma cell line HepG2. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the RXRα (lanes 3 and 4), PPARα (lanes 6 and 7), and FXRα (lanes 9 and 10) expression vectors as indicated. In addition, cells were treated with 1 μM retinoic acid (RA; lanes 2 and 4), 1 mM clofibric acid (CA; lanes 5 and 7), and 100 mM chenodeoxycholic acid (CDCA; lanes 8 and 10) to activate the RXRα, PPARα, and FXRα nuclear receptors, respectively. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates (RI) relative to that of the HBV DNA (4.1-kbp) construct in the absence of nuclear receptor expression or ligand (left bars) are reported. The mean RNA and DNA levels plus standard deviations from four independent analyses are shown. The levels of the transcripts and replication intermediates in the nuclear receptor and/or ligand-treated cells which are statistically significantly higher than their levels in the corresponding untreated cells by Student's t test (P < 0.05) are indicated with asterisks.
