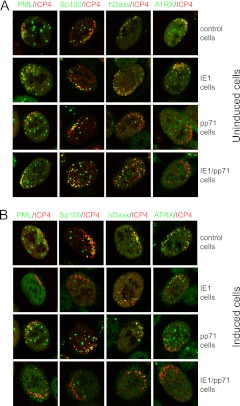
Fig 9.
Immunofluorescence analysis of the recruitment of ND10 components to sites associated with HSV-1 genomes in control, IE1, pp71, and IE1/pp71 cells. Cells were infected with dl1403/CMVlacZ and then stained for ICP4 and the indicated ND10 proteins at 24 h after infection. (A) The infections were performed in uninduced cells at an MOI of 0.5, 0.2, 0.05, and 0.05 for control, IE1, pp71, and IE1/pp71 cells, respectively. The MOI was varied between cell lines to ensure a convenient number of plaques for analysis. (B) The infections were performed after induction with 100 ng/ml doxycycline for 24 h, using an MOI of 0.5, 0.01, 0.02, and 0.005 for control, IE1, pp71, and IE1/pp71 cells, respectively. Samples were stained as indicated for ICP4 (MAb 58S) and rabbit antibodies for PML, Sp100, hDaxx, and ATRX. Secondary antibodies were anti-mouse Alexa 555 and anti-rabbit Alexa 633 (shown in the green channel here). Cells at the periphery of developing plaques with asymmetric distributions of HSV-1 genomes and early replication compartments were identified visually and then scanned to determine the distributions of the ND10 components. The quantitative data given in the text were determined by examination of 20 cells in each case.