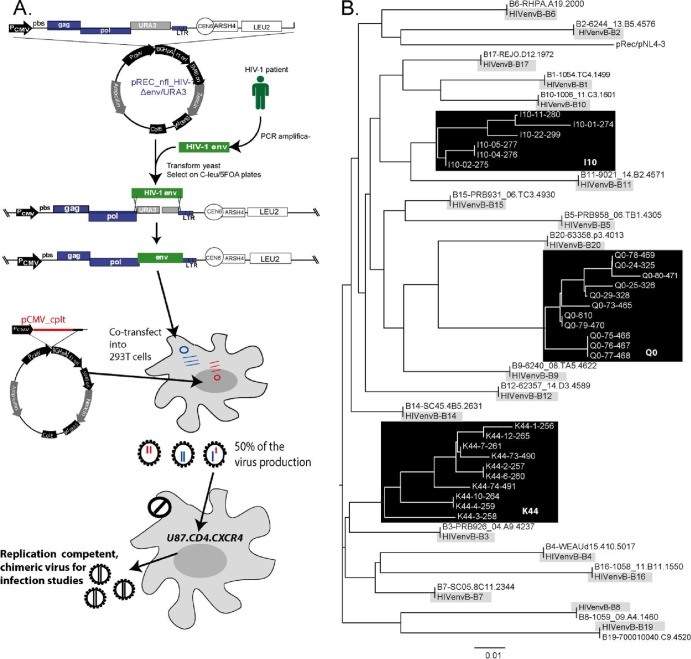
Fig 1.
Construction and sequence analyses of the HIV-1 env chimeric viruses derived from acute and chronic infection samples. (A) An HIV-1 env cassette was reverse transcription-PCR (RT-PCR) amplified as described in Materials and Methods and then transfected into yeast along with the linearized pREC_nfl_HIV-1Δenv/URA3. The URA3 gene is replaced by the env cassette via homologous recombination/gap repair, and this process is selected via yeast colony growth on plates lacking leucine but containing FOA. FOA is converted into a toxic anabolite by the uracil biosynthesis machinery encoded by URA3. The pREC_nfl_HIV-1 vectors with chronic or acute env genes are then purified and used to transfect 293T cells along with the complementing vector, pCMV_cplt by the method of Dudley et al. (17). Virus-containing supernatant from these transfections are propagated for a maximum of 2 weeks on U87.CD4.CCR5 cells to produce the virus employed in all subsequent assays. Abbreviations: PCMV, promoter for cytomegalovirus: pbs, primer-binding sequence; LTR, long terminal repeat; PCR amplifica-, PCR amplification. (B) The viruses amplified on the U87.CD4.CCR5 cells served as the templates for RT-PCR and DNA sequencing. The neighbor-joining phylogenetic tree was constructed based on the C2-V3 region of env within the propagated viruses. For each of the acute clones, the original HIV-1 env gene derived from SGA of the patient sample is provided to verify sequence identity to the cloned virus.