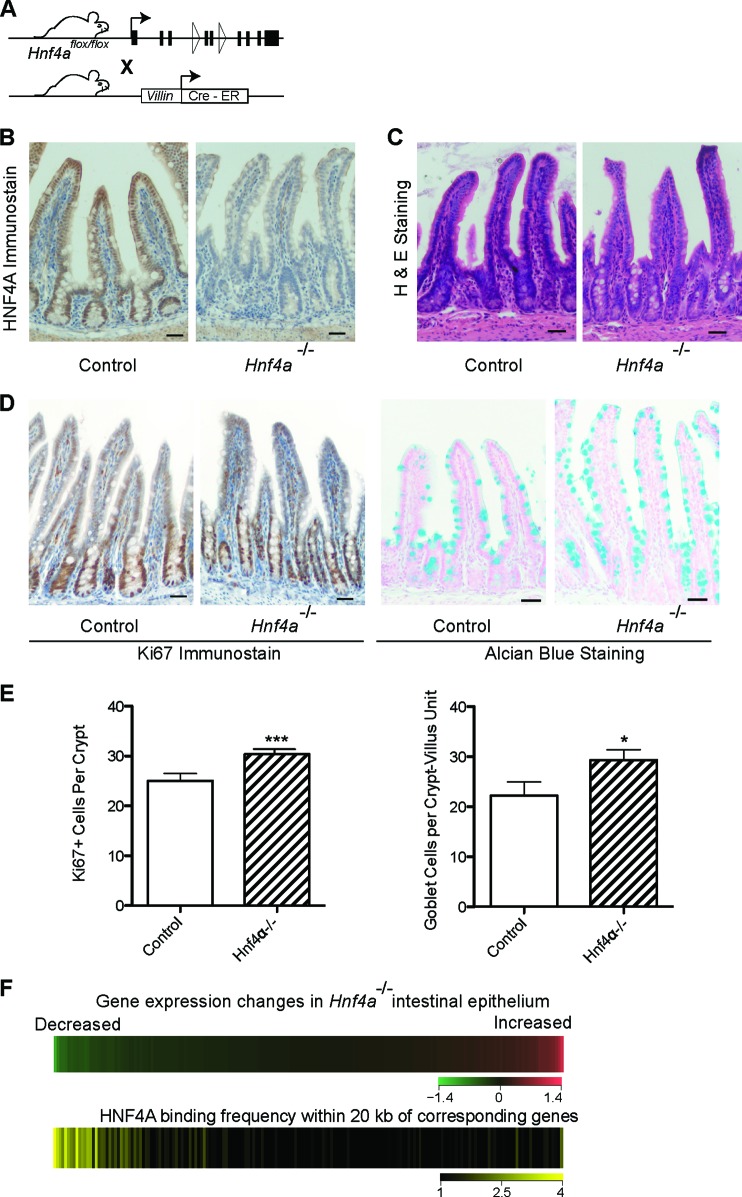
Fig 2.
HNF4A functions as a transcriptional activator in intestinal epithelium. (A) Schema for inducible, intestine-specific depletion of HNF4A in vivo. (B) Immunostaining verifies efficient HNF4A loss in intestinal epithelium after tamoxifen-induced Cre recombination. (C) Hematoxylin and eosin staining shows that Hnf4a−/− gut epithelium is overtly intact. (D) The proportions of Ki67+ proliferating crypt cells and Alcian blue-avid goblet cells are slightly increased in Hnf4a−/− gut epithelium, as others have reported. Scale bar, 30 μm. (E) The small but significant increase in Ki67+ and goblet cells was quantified by counting labeled cells (data represent the mean cell counts ± standard deviations from >3 mice; *, P < 0.05; ***, P < 0.001). (F) HNF4A activates transcription in intestinal villi. We partitioned all transcripts, measured by microarray analysis (top heat map), in bins of 100, from those most reduced in Hnf4a−/− intestines (green) to those most increased (red) and calculated the HNF4A binding frequency near these groups of 100 genes in ChIP-Seq analysis of WT intestines (bottom heat map). HNF4A binding was highest near genes whose levels drop with HNF4A loss.