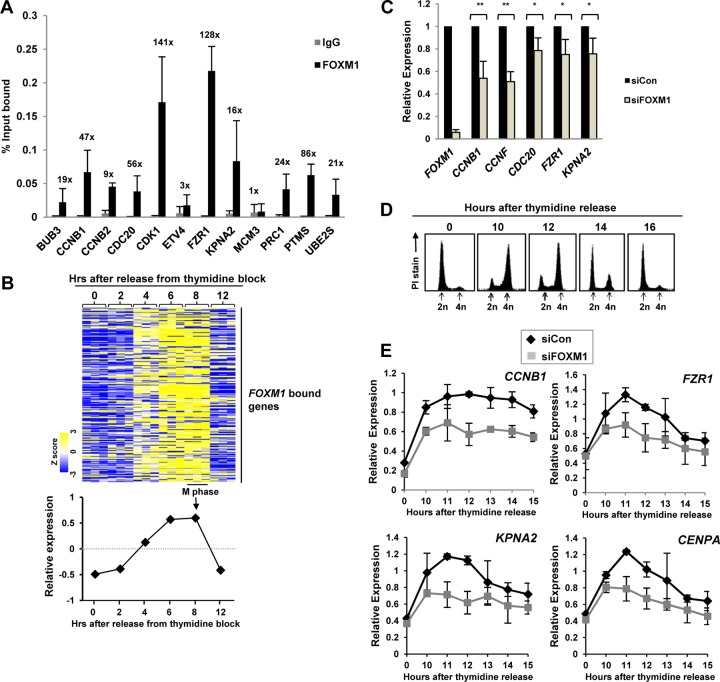
Fig 2.
FOXM1 directly activates late cell cycle genes. (A) ChIP-qPCR validation of FOXM1 binding to regions associated with the indicated genes. Experiments were carried out with three biological repeats. Bars indicate the average percentages of input precipitated with the FOXM1 antibody or nonspecific IgG with the standard deviations. The numbers above each set of bars represent the fold increase in signal with FOXM1 over IgG. (B) Heat map from an expression microarray analysis performed in HeLa cells after release from a double thymidine block (25) of the 270 genes that are associated with FOXM1 binding regions. The graph below summarizes the average Z score of the expression profiles of this group of genes. (C and E) RT-qPCR analysis of the expression of the indicated genes in U2OS cells grown asynchronously (C) or subjected to a double thymidine block and released for the indicated times (E). Cells were pretreated with a nontargeting siRNA (siCon) or siRNA against FOXM1. The data are the averages of two (E) or three (C) experiments and are shown for each gene relative to its expression in the presence of the control siRNA (taken as 1 [C]) or the internal control gene HMBS (E). ** and * represent P values of <0.01 and <0.05, respectively. (D) DNA content profiles of U2OS cells at the indicated time points following release from a double thymidine block. DNA content is determined by propidium iodide (PI) staining, and peaks corresponding to cells before (2n) and after (4n) DNA replication are shown.