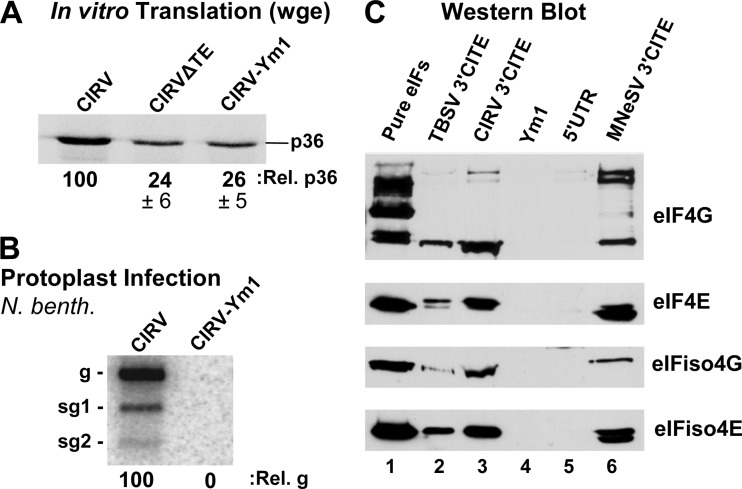
Fig 3.
Interaction of the CIRV 3′CITE with eIF4F and eIFiso4F. (A) SDS-PAGE analysis of p36 production in wge. CIRV-ΔTE is a 3′-truncated version of CIRV that does not contain a 3′CITE; CIRV-Ym1 contains a mutated 3′CITE (mutation shown in Fig. 1B). (B) Northern blot showing genomic (g) and subgenomic (sg1 and sg2) viral RNA isolated from cucumber protoplasts 22 h after inoculation with the genomic RNAs indicated above the blot. N. benth, N. benthamiana. (C) Western blots showing the detection of the eIFs indicated to the right. Blots were performed on eluates from streptomycin-conjugated Sepharose columns containing StreptoTagged versions of the RNAs indicated above. Prior to elution, columns loaded with RNA were incubated with wge and then washed. Lane 1 contained 2 pmol of each purified eIF; lanes 2 and 3 contained eluate from the wt Y-shaped 3′CITEs of TBSV and CIRV, respectively; lane 4 contained eluate from the CIRV 3′CITE mutant Ym1; lane 5 contained eluate from RNA corresponding to the CIRV 5′UTR; lane 6 contained eluate from the I-shaped 3′CITE of MNeSV, which was used as a positive control.