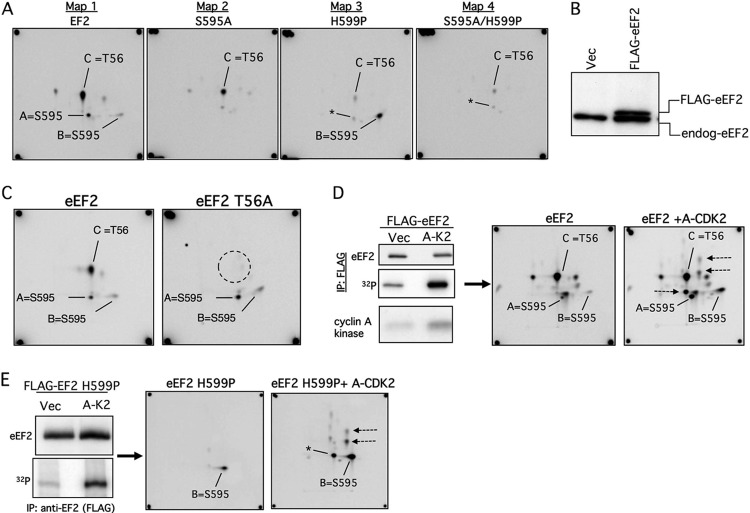
Fig 3.
eEF2 S595 phosphorylation in vivo. (A) Phosphopeptide maps of eEF2 (map 1) and the indicated mutants (maps 2 to 4) isolated from 293T cells labeled with [32P]orthophosphate. The peptides representing S595 (spots A and B) and T56 (spot C) are indicated. The S595A mutation abrogates the S595 spots in both the WT and H599P backgrounds (maps 2 and 4). Note that the amount of T56 phosphorylation is reduced in the S595A and H599P mutants (maps 2 to 4). Asterisks indicate a minor spot that comigrates with spot A. (B) Amount of overexpression of transfected FLAG-eEF2 relative to endogenous eEF2 (endog-eEF2). Vec, vector. (C) Identification of spot C as containing phosphorylated T56. Cells transfected with WT eEF2 or eEF2 T56A were labeled with orthophosphate and the immunoprecipitated eEF2 proteins analyzed by phosphopeptide mapping. (D) eEF2 phosphorylation is increased by cyclin A-CDK2 expression. U2OS cells were transfected with eEF2 and either cyclin A-CDK2 or empty vector (Vec), and eEF2 was immunoprecipitated after [32P]orthophosphate labeling (top two panels). The bottom panel shows total cyclin A-CDK histone H1 kinase activity. Phosphopeptide mapping reveals increased phosphorylation of S595 (spots A and B) and other sites (arrows). (E) Analysis similar to that described for panel D using eEF2 H599P.