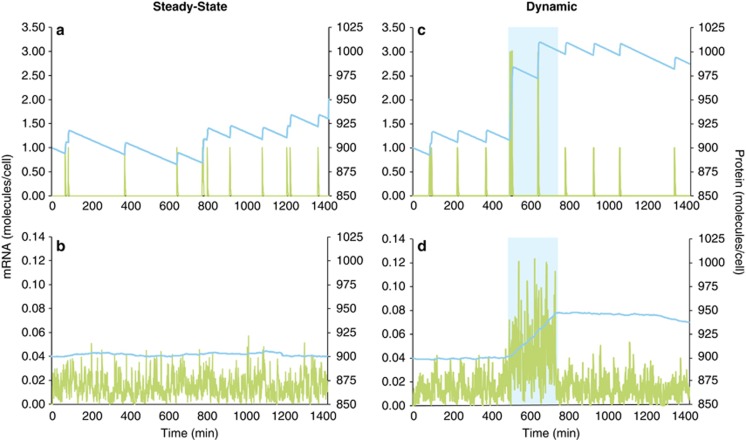
Figure 3.
Simulation model of levels of mRNA (green lines) and protein (blue lines) of the same gene in a single cell (a, c) or averaged for a population of 100 cells (b, d) during a 24-h period. In the steady-state version of the model (a, b), which could represent either constitutive gene expression or an unchanging extrinsic regulatory signal, each cell experiences up to 10 randomly timed transcription events per day and produces a single mRNA molecule at each event (Supplementary Materials). In the dynamic version (c, d), extrinsic signaling upregulates gene transcription for a 4-h period (500–740 min; blue shading) through an increase in transcriptional burst size to three mRNA molecules per transcription event. Both model types were initialized with 900 protein molecules per cell, and both assume 7 proteins are translated from each mRNA template, that the half-life of mRNA is 1.5 min, and that the half-life of protein is 12 h. Varying the parameter values (for example, frequency of transcription events, mRNA burst size, proteins translated per message, macromolecule half-lives) changes the size of the final mRNA and protein pools, but they remain poorly synchronized under dynamic extrinsic conditions.