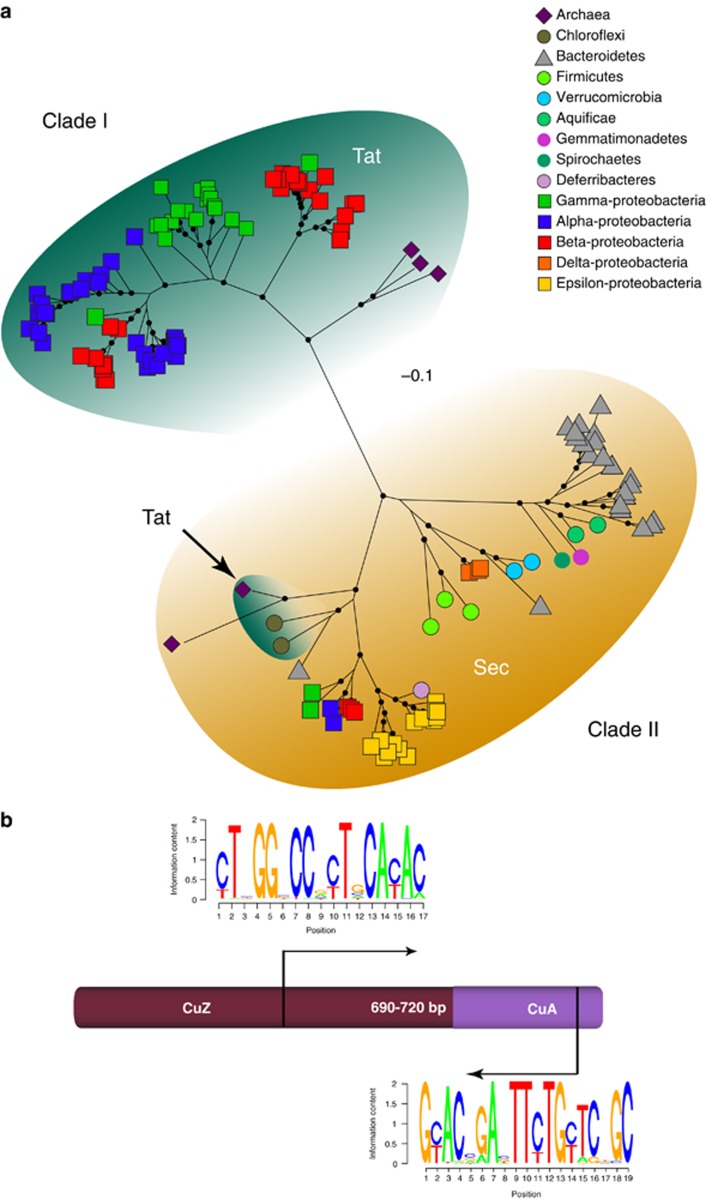
Figure 1.
(a) Unrooted maximum likelihood phylogeny of full-length nosZ amino-acid sequences obtained from genomes. The distribution of signal peptide motif detected for each clade is indicated. Symbols on tree tips specify major taxonomic groups, and scale bar indicates corrected substitutions per site (LG+Γ+F for amino acids; Binary+Γ for indel sites). Nodes with >70% bootstrap support (n=500) are denoted by dots. (b) Schematic representation of nosZ gene indicating the center multinuclear copper catalytic site (CuZ) and the C-terminal cupredoxin active site (CuA). Arrows indicate primer binding sites and amplicon size, and the consensus sequence of clade II for each site is shown as SeqLogo diagrams.